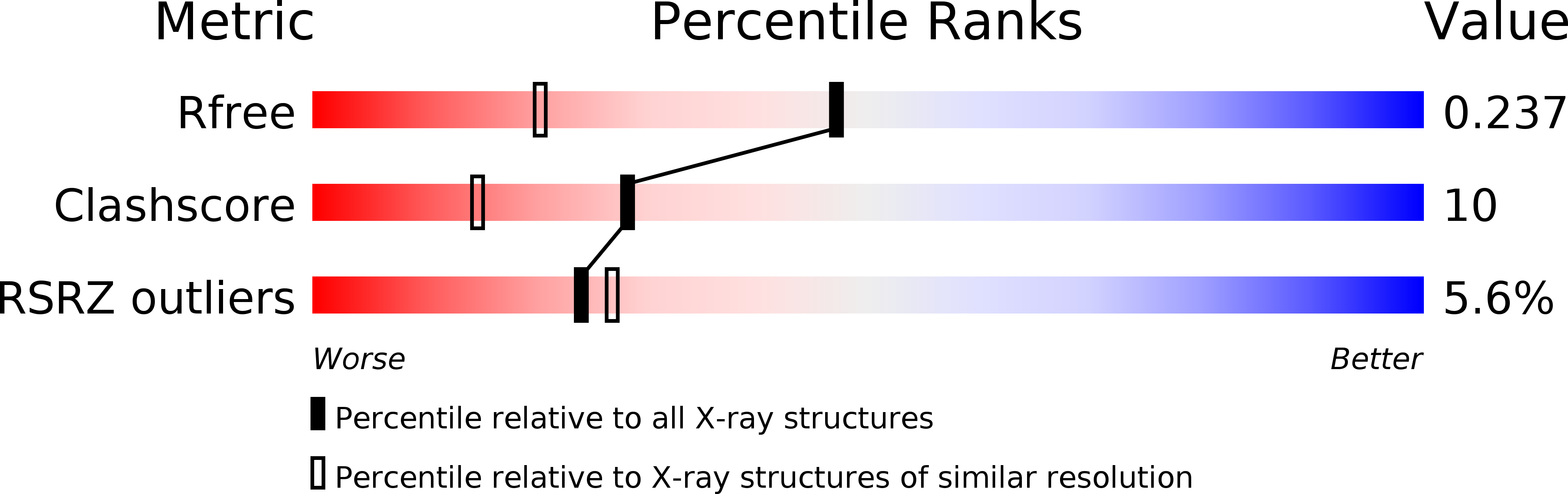Deposition Date
2004-09-26
Release Date
2005-03-15
Last Version Date
2024-04-03
Entry Detail
PDB ID:
1XJX
Keywords:
Title:
The crystal structures of the DNA binding sites of the RUNX1 transcription factor
Biological Source:
Source Organism:
Method Details:
Experimental Method:
Resolution:
1.70 Å
R-Value Free:
0.25
R-Value Work:
0.16
R-Value Observed:
0.16
Space Group:
P 43