Deposition Date
2004-09-24
Release Date
2005-02-01
Last Version Date
2024-02-14
Entry Detail
PDB ID:
1XJR
Keywords:
Title:
The Structure of a Rigorously Conserved RNA Element Within the SARS Virus Genome
Method Details:
Experimental Method:
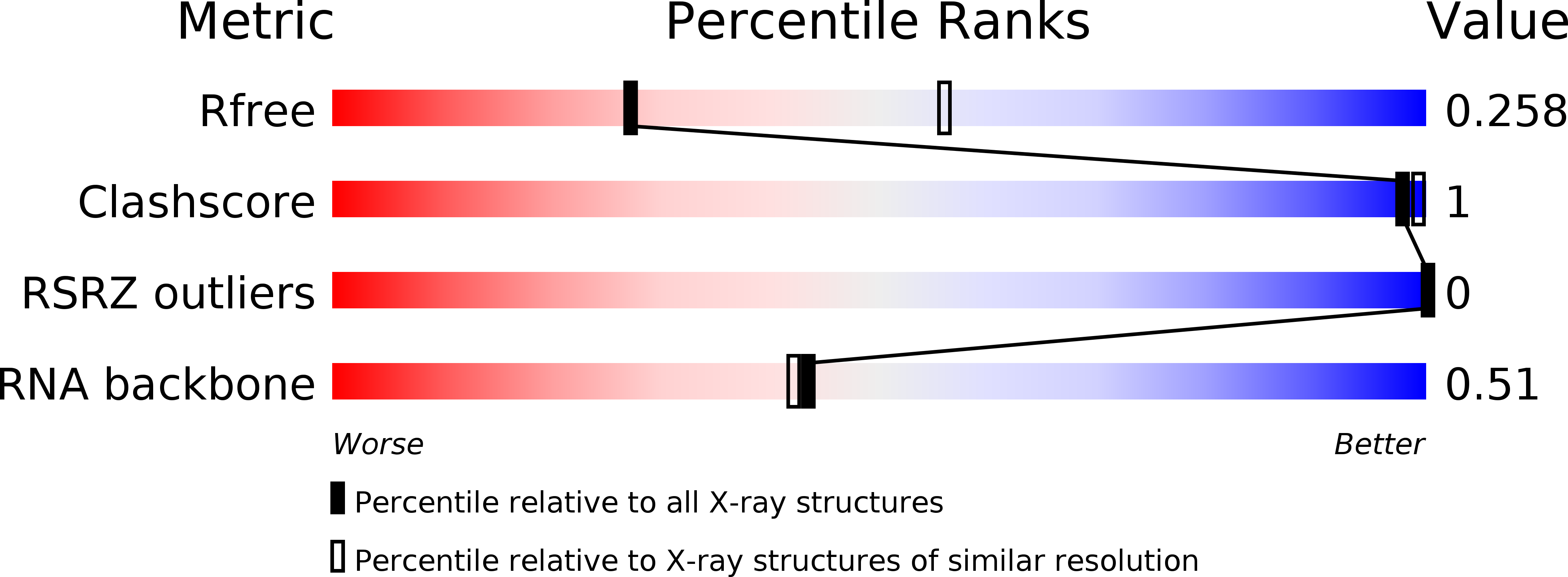
Resolution:
2.70 Å
R-Value Free:
0.24
R-Value Work:
0.23
R-Value Observed:
0.23
Space Group:
P 65 2 2