Deposition Date
1988-02-24
Release Date
1988-04-16
Last Version Date
2024-02-14
Entry Detail
PDB ID:
1XIA
Keywords:
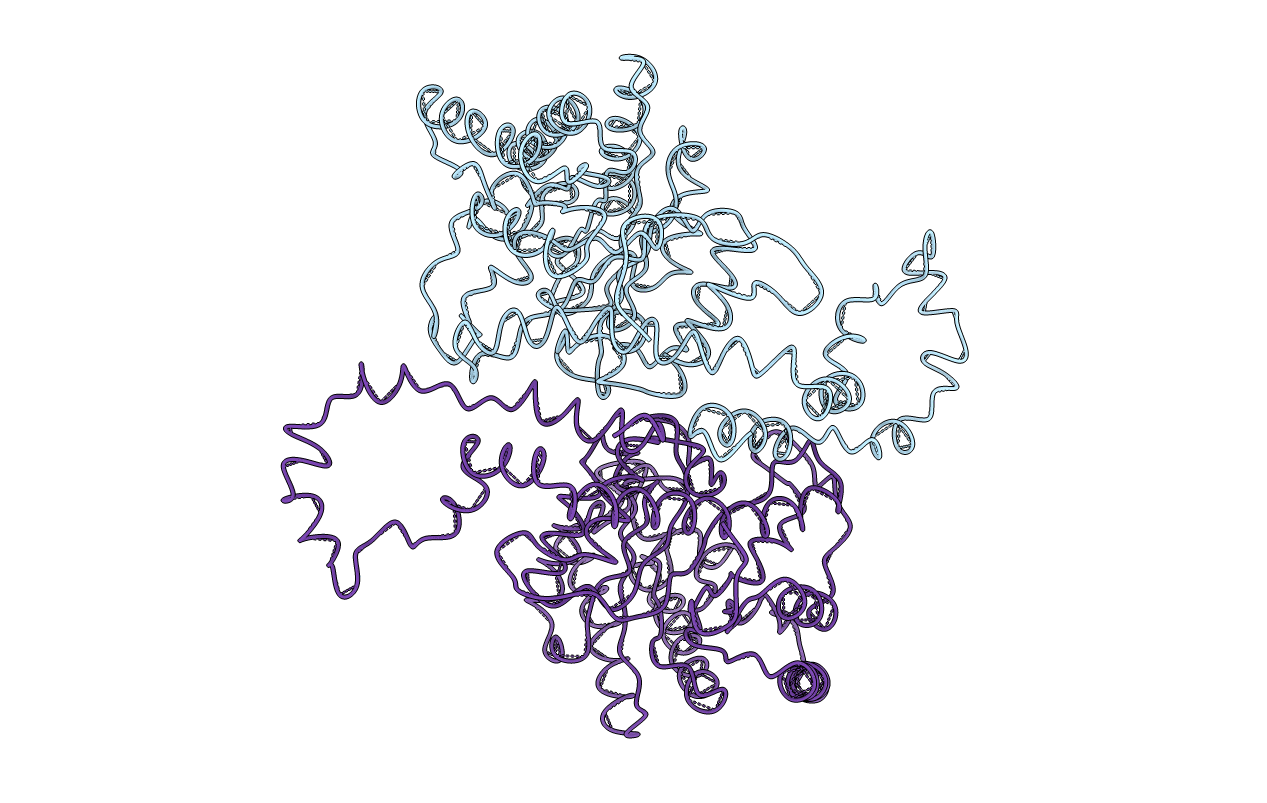
Title:
COMPARISON OF BACKBONE STRUCTURES OF GLUCOSE ISOMERASE FROM STREPTOMYCES AND ARTHROBACTER
Biological Source:
Source Organism(s):
Arthrobacter sp. NRRL (Taxon ID: 1669)
Method Details: