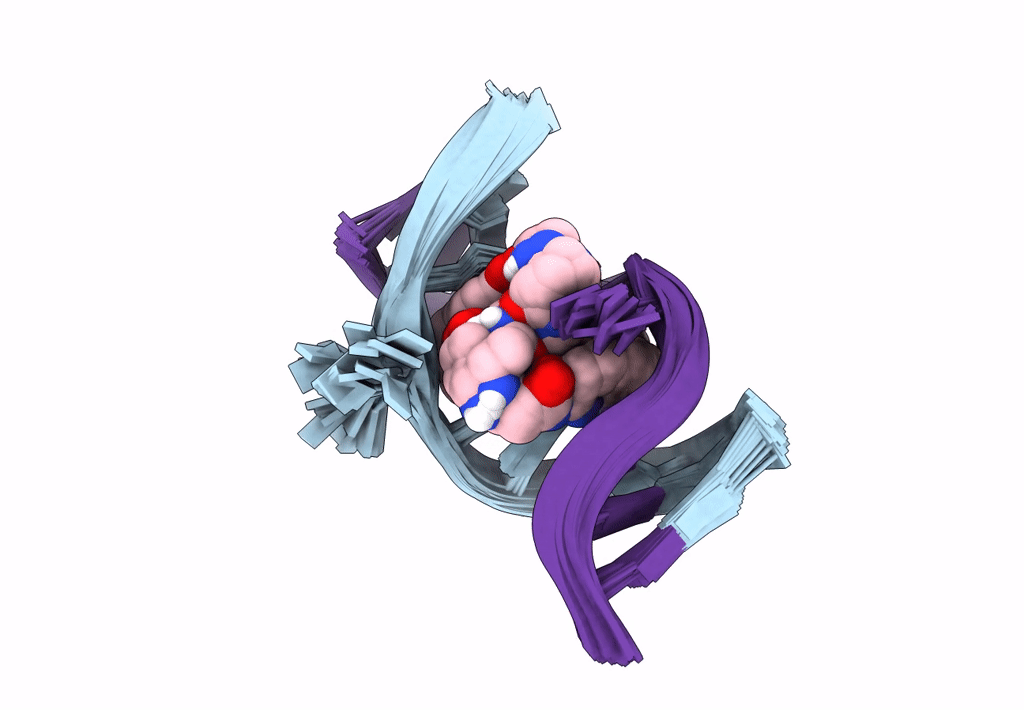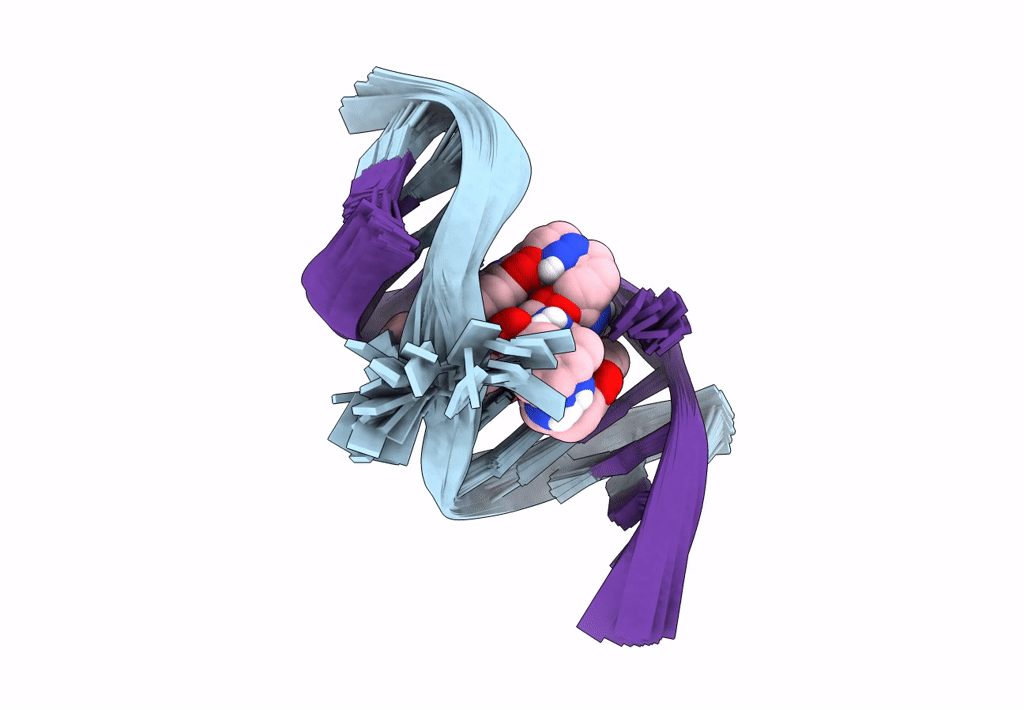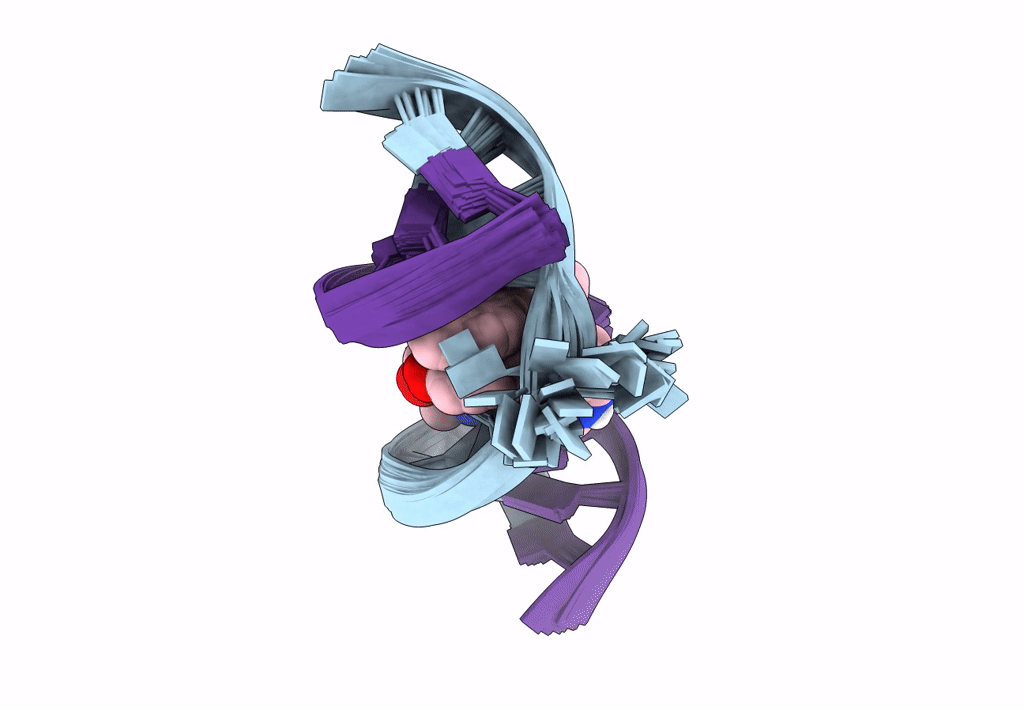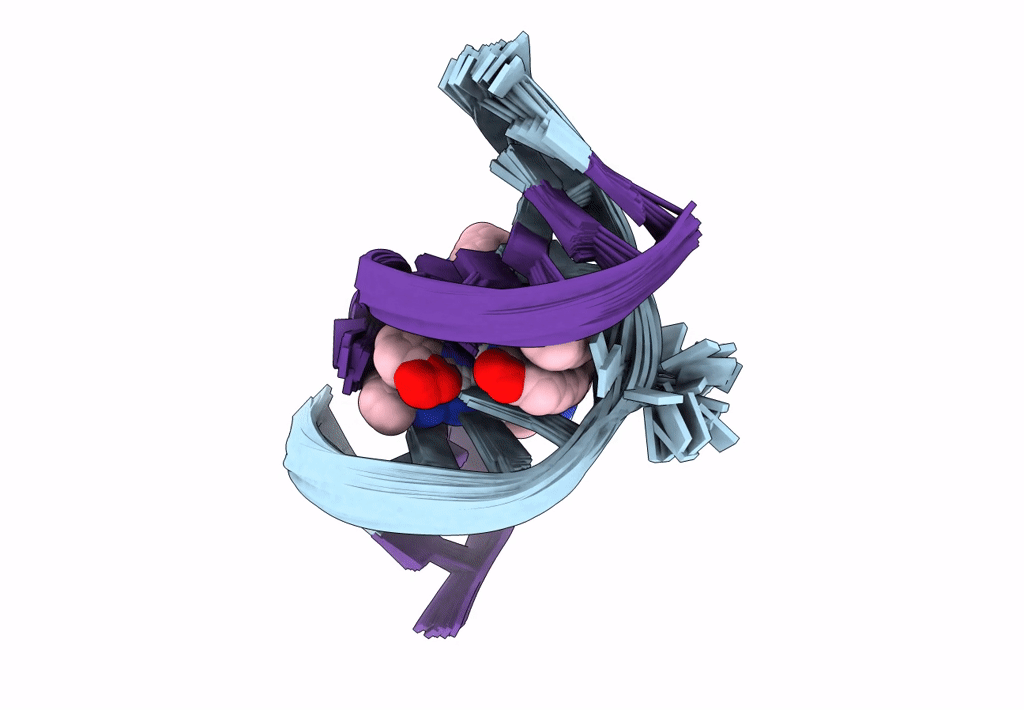
Deposition Date
2005-04-20
Release Date
2006-04-04
Last Version Date
2024-05-29
Entry Detail
PDB ID:
1X26
Keywords:
Title:
Solution structure of the AA-mismatch DNA complexed with naphthyridine-azaquinolone
Method Details:
Experimental Method:
Conformers Calculated:
150
Conformers Submitted:
30
Selection Criteria:
structures with the lowest energy


