Deposition Date
2004-10-25
Release Date
2005-06-27
Last Version Date
2024-05-15
Entry Detail
PDB ID:
1WA8
Keywords:
Title:
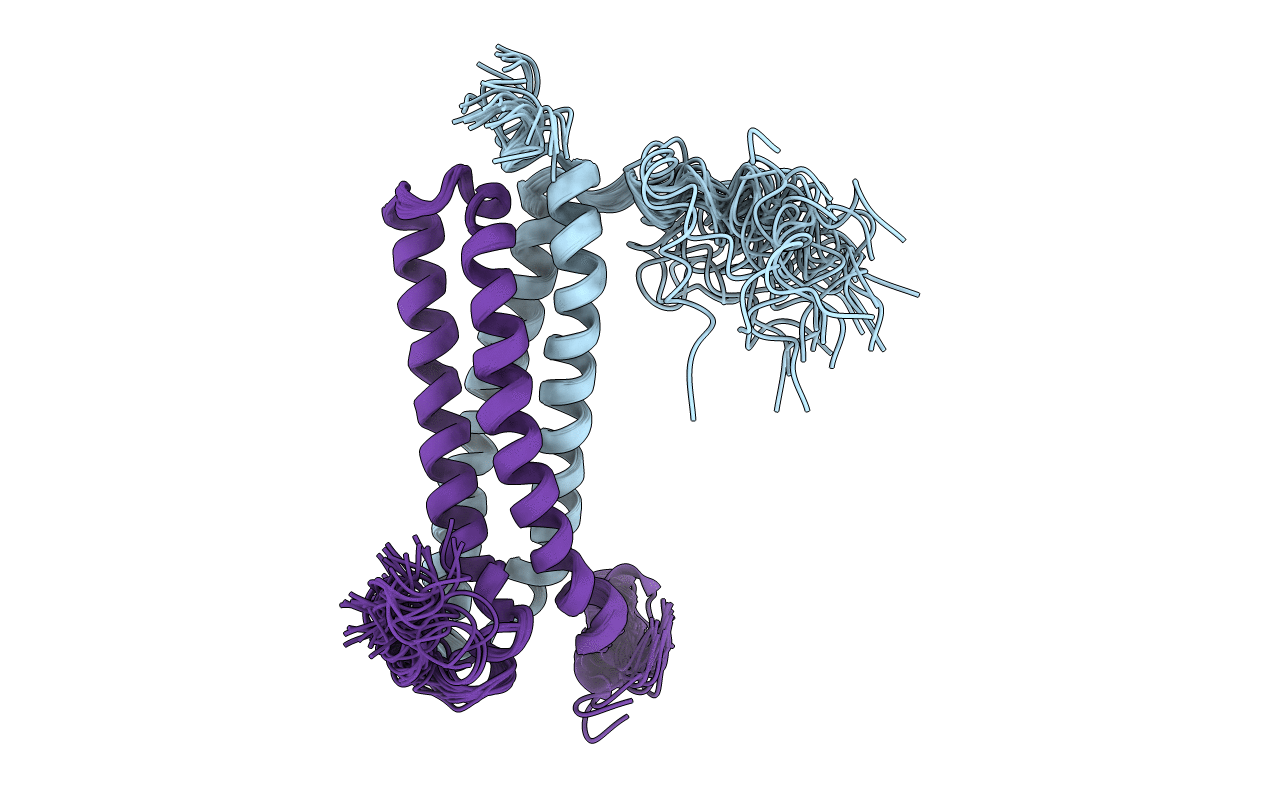
Solution Structure of the CFP-10.ESAT-6 Complex. Major Virulence Determinants of Pathogenic Mycobacteria
Biological Source:
Source Organism(s):
MYCOBACTERIUM BOVIS (Taxon ID: 1765)
MYCOBACTERIUM TUBERCULOSIS (Taxon ID: 83332)
MYCOBACTERIUM TUBERCULOSIS (Taxon ID: 83332)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
100
Conformers Submitted:
28
Selection Criteria:
LEAST RESTRAINT VIOLATION