Deposition Date
2004-09-23
Release Date
2005-03-01
Last Version Date
2024-11-13
Entry Detail
PDB ID:
1W8K
Keywords:
Title:
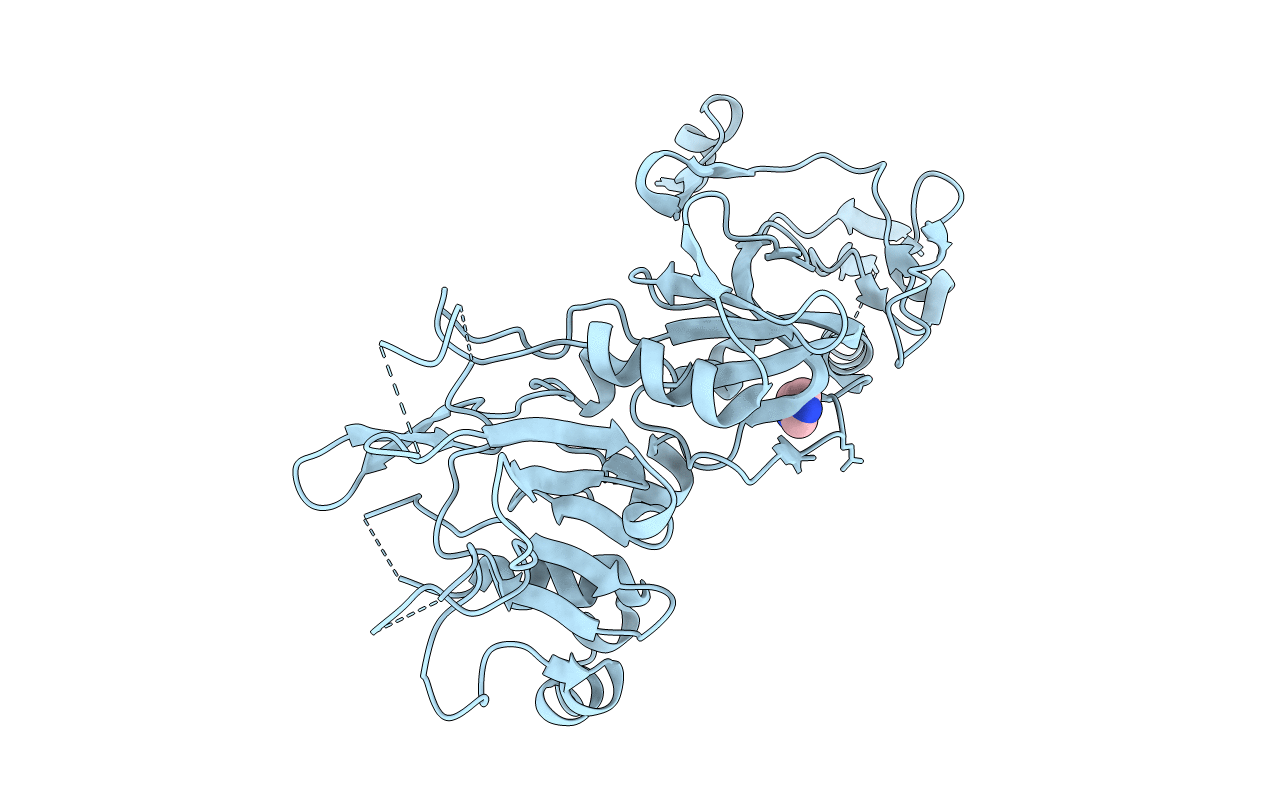
Crystal structure of apical membrane antigen 1 from Plasmodium vivax
Biological Source:
Source Organism(s):
PLASMODIUM VIVAX (Taxon ID: 5855)
Expression System(s):
Method Details:
Experimental Method:
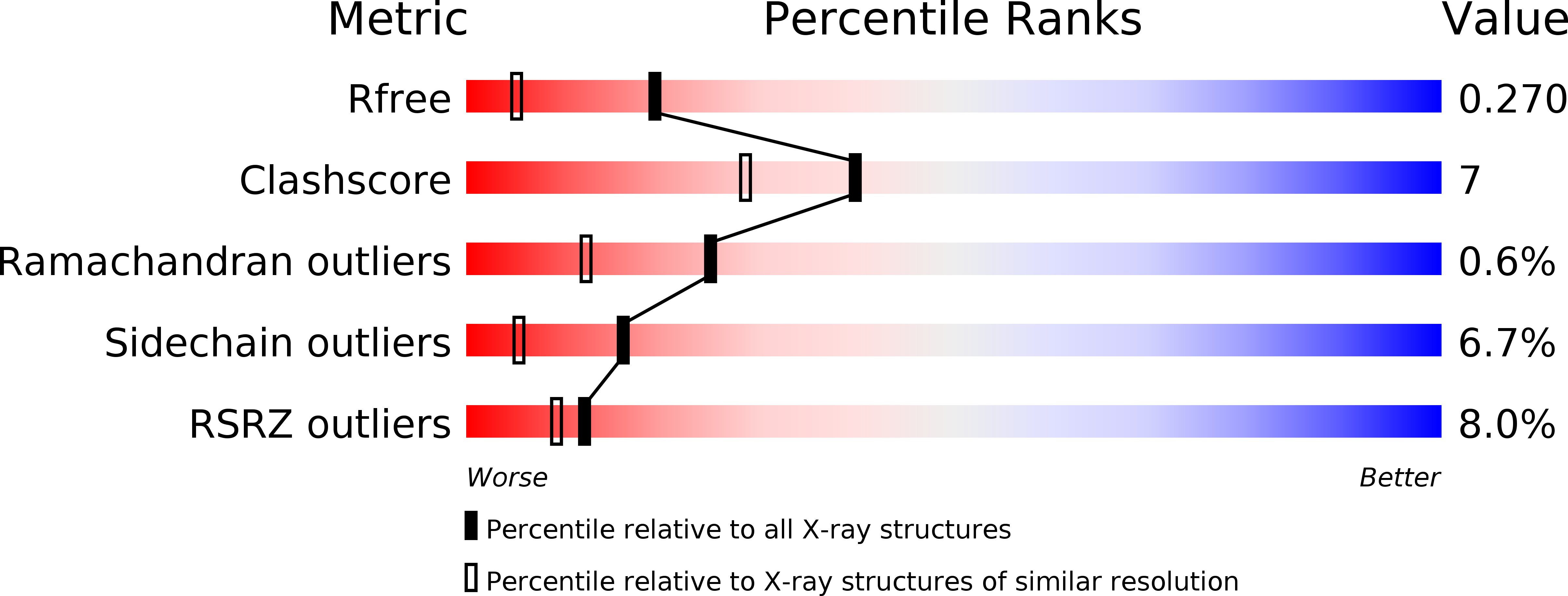
Resolution:
1.80 Å
R-Value Free:
0.26
R-Value Work:
0.21
R-Value Observed:
0.22
Space Group:
C 1 2 1