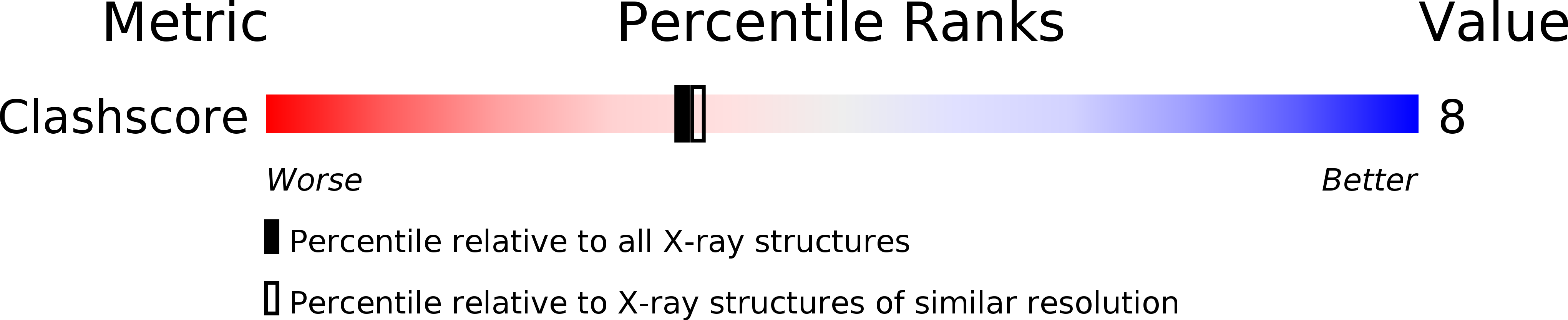
Deposition Date
1988-08-18
Release Date
2011-07-13
Last Version Date
2023-12-27
Entry Detail
PDB ID:
1VT5
Keywords:
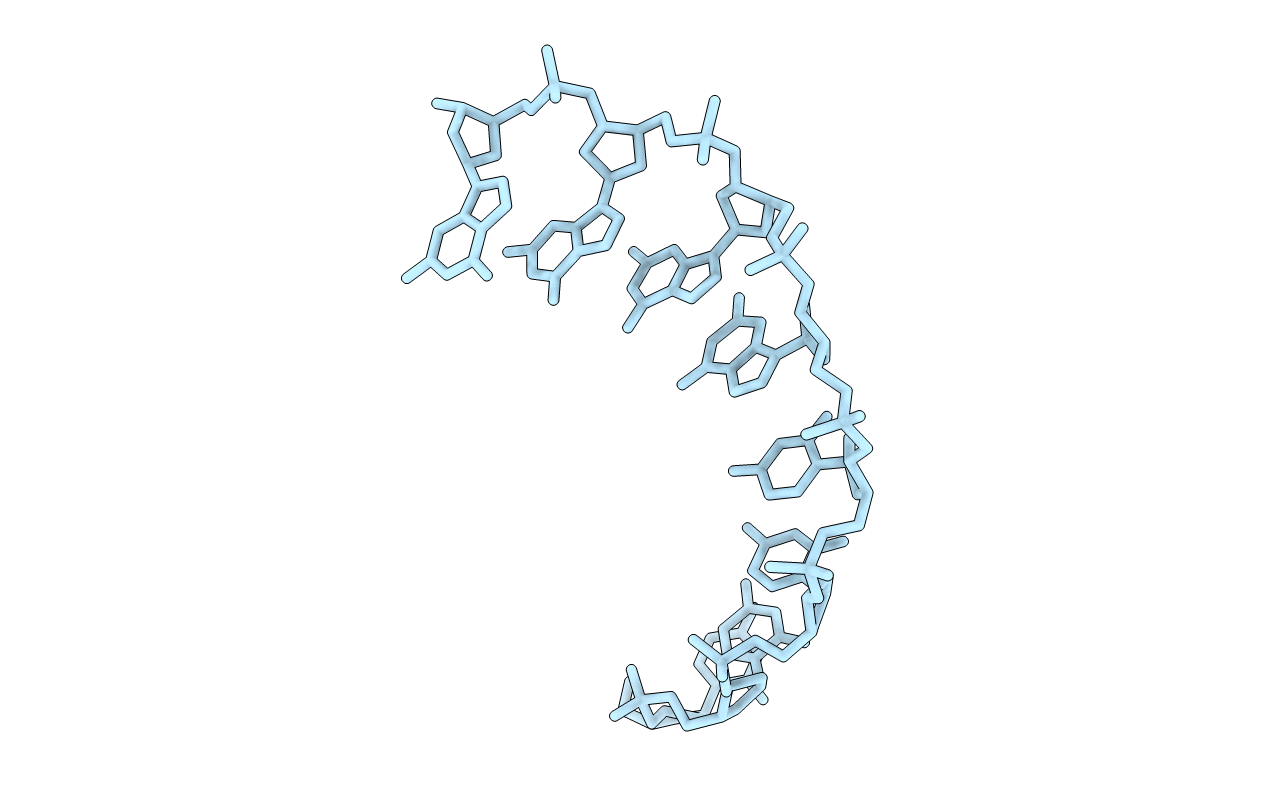
Title:
THE CRYSTAL STRUCTURE OF D(CCCCGGGG): A NEW A-FORM VARIANT WITH AN EXTENDED BACKBONE CONFORMATION
Biological Source:
Source Organism:
Method Details:
Experimental Method:
Resolution:
2.25 Å
R-Value Work:
0.24
R-Value Observed:
0.24
Space Group:
P 43 21 2