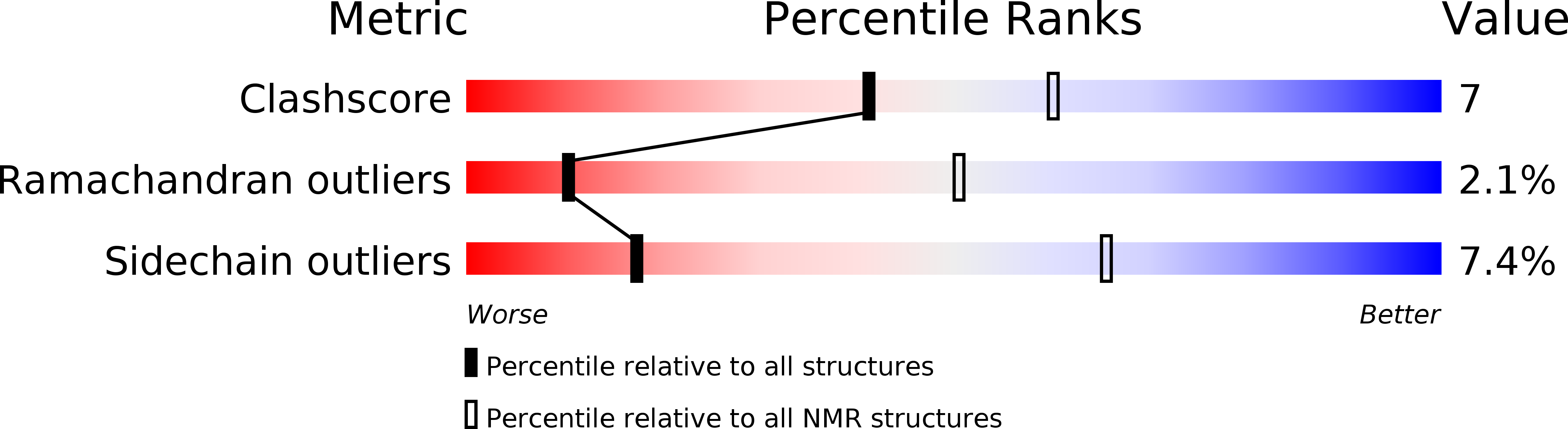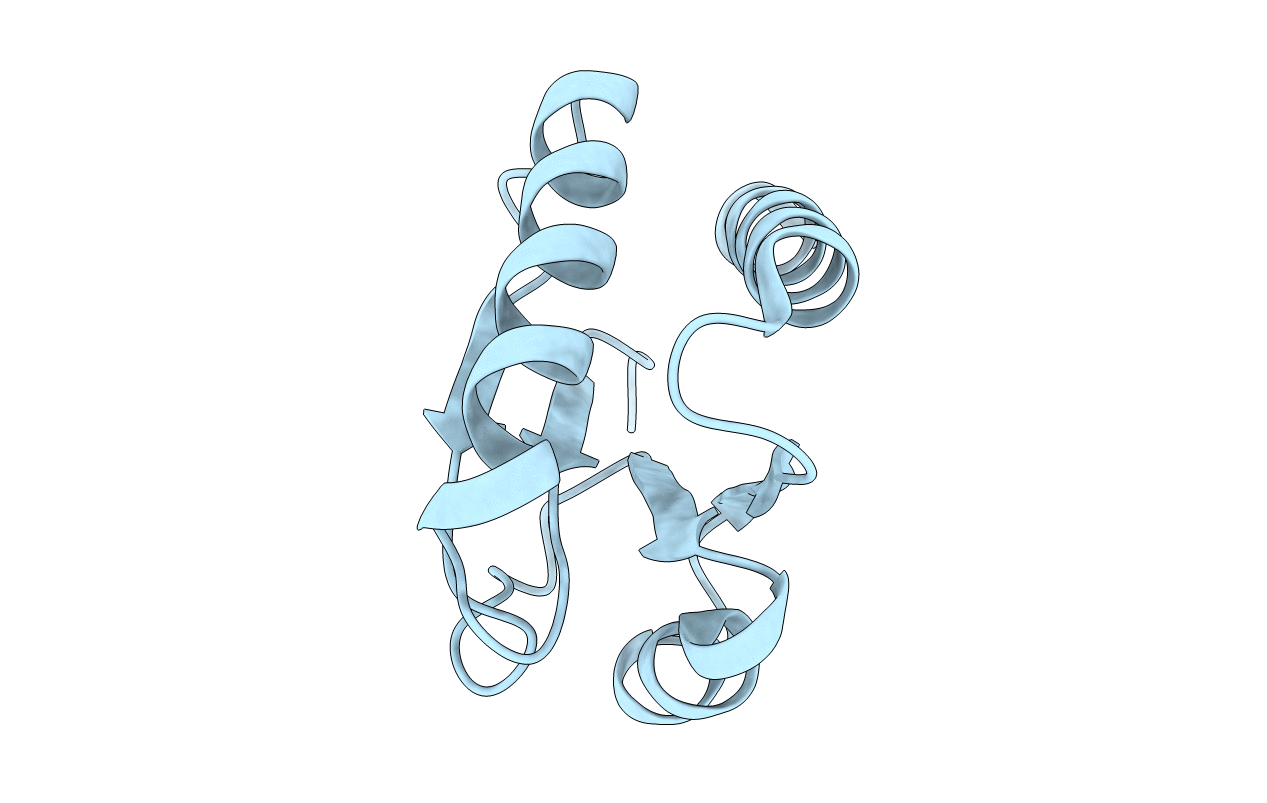
Deposition Date
2004-06-14
Release Date
2004-09-21
Last Version Date
2023-12-27
Entry Detail
PDB ID:
1VKR
Keywords:
Title:
STRUCTURE OF IIB DOMAIN OF THE MANNITOL-SPECIFIC PERMEASE ENZYME II
Biological Source:
Source Organism(s):
Escherichia coli (Taxon ID: 83334)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
80
Conformers Submitted:
1
Selection Criteria:
REGULARIZED MEAN STRUCTURE