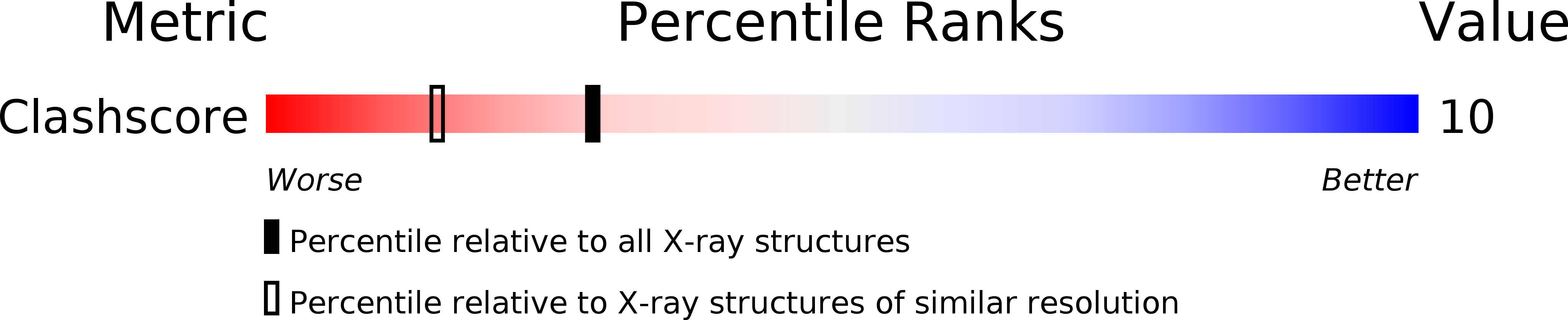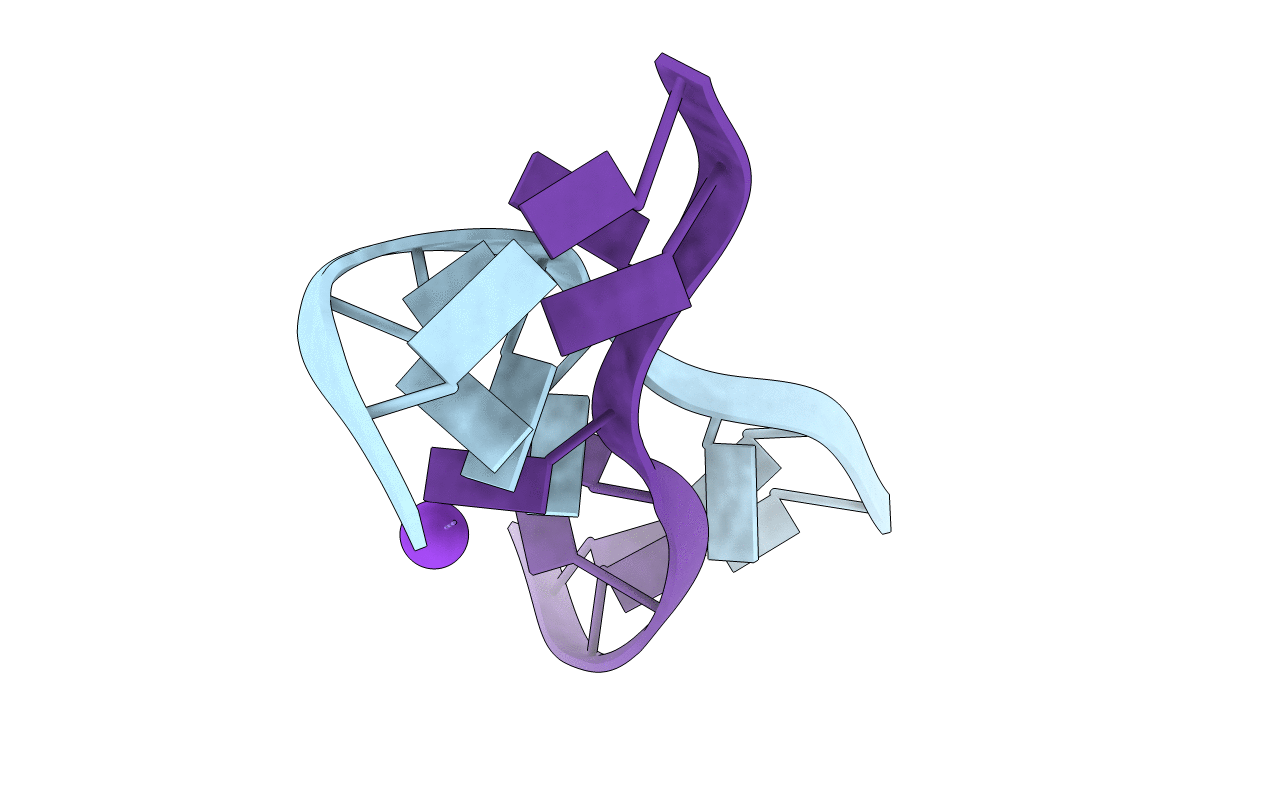
Deposition Date
2003-11-03
Release Date
2004-06-08
Last Version Date
2023-10-25
Entry Detail
PDB ID:
1V3O
Keywords:
Title:
Crystal structure of d(GCGAGAGC): the DNA quadruplex structure split from the octaplex
Method Details:
Experimental Method:
Resolution:
1.70 Å
R-Value Free:
0.29
R-Value Work:
0.26
Space Group:
I 2 2 2