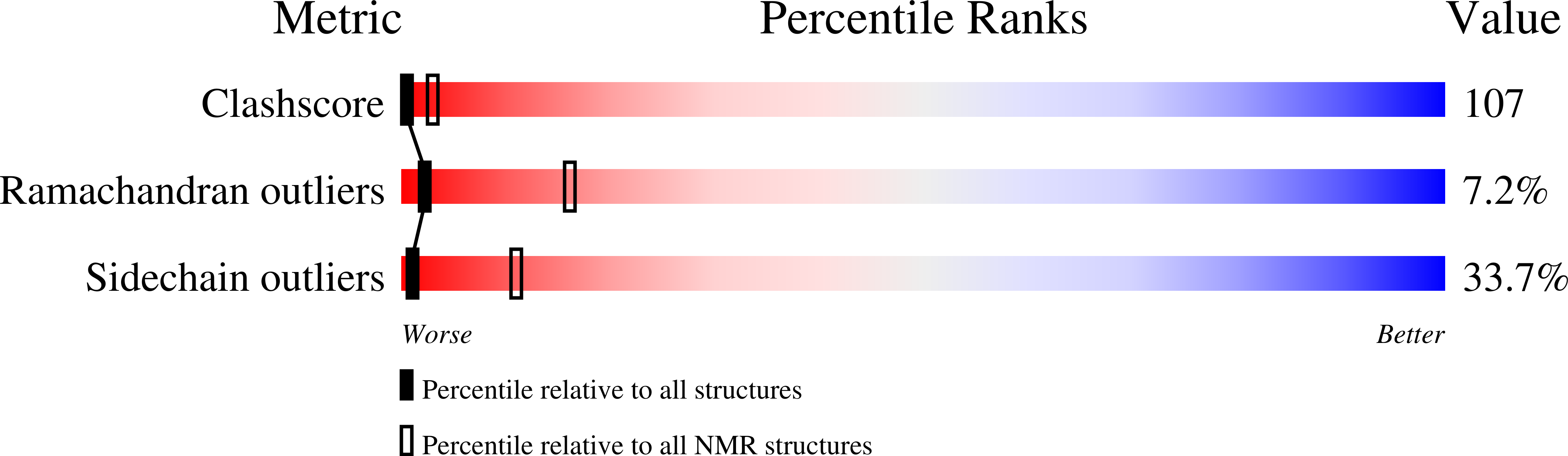
Deposition Date
1997-12-05
Release Date
1998-06-10
Last Version Date
2024-05-22
Entry Detail
PDB ID:
1UWO
Keywords:
Title:
CALCIUM FORM OF HUMAN S100B, NMR, 20 STRUCTURES
Biological Source:
Source Organism:
Homo sapiens (Taxon ID: 9606)
Host Organism:
Method Details:
Experimental Method:
Conformers Calculated:
30
Conformers Submitted:
20
Selection Criteria:
LEAST RESTRAINT VIOLATIONS