Deposition Date
2004-01-27
Release Date
2004-08-05
Last Version Date
2024-05-15
Entry Detail
PDB ID:
1UW0
Keywords:
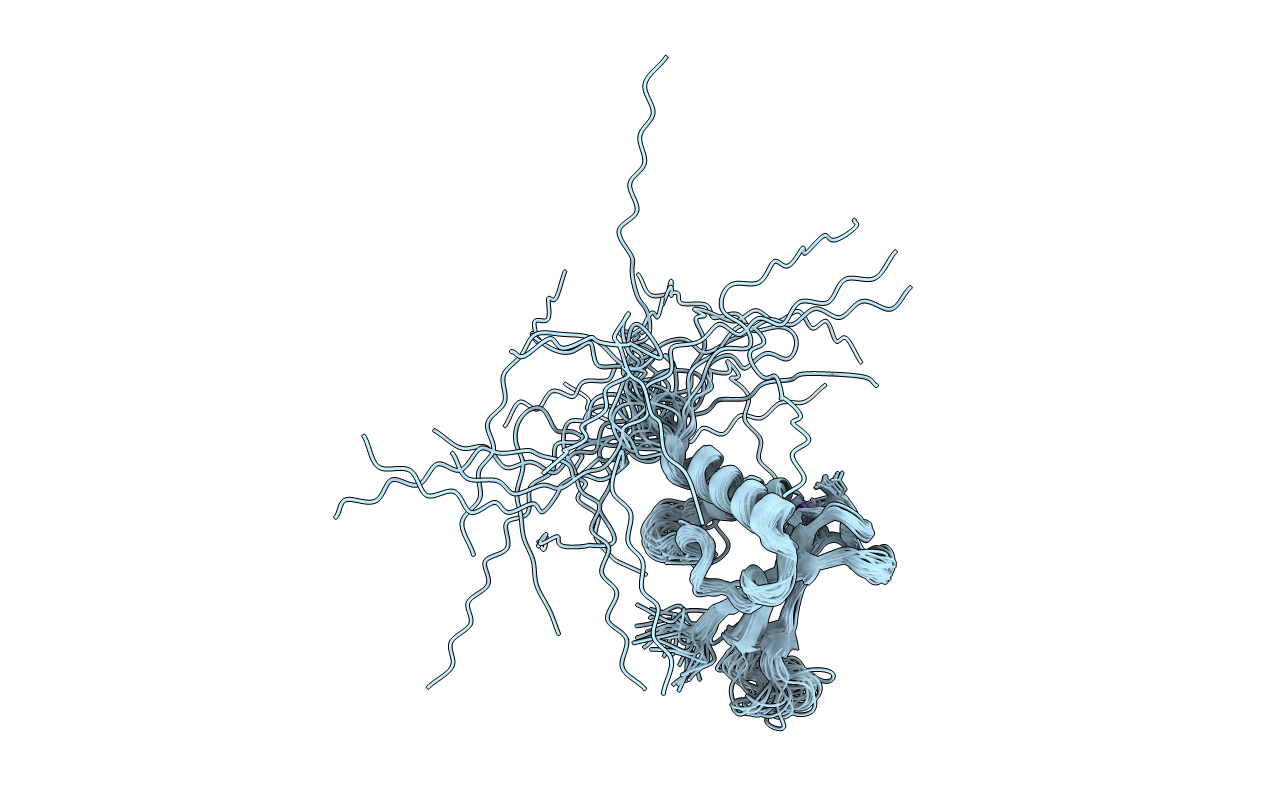
Title:
Solution structure of the zinc-finger domain from DNA ligase IIIa
Biological Source:
Source Organism:
HOMO SAPIENS (Taxon ID: 9606)
Host Organism:
Method Details:
Experimental Method:
Conformers Calculated:
50
Conformers Submitted:
28
Selection Criteria:
LOW RESTRAINT VIOLATIONS


