Deposition Date
2004-01-21
Release Date
2004-02-19
Last Version Date
2025-12-17
Entry Detail
PDB ID:
1UVK
Keywords:
Title:
The structural basis for RNA specificity and Ca2 inhibition of an RNA-dependent RNA polymerase phi6p2 dead-end complex
Biological Source:
Source Organism(s):
Cystovirus phi6 (Taxon ID: 10879)
synthetic construct (Taxon ID: 32630)
synthetic construct (Taxon ID: 32630)
Expression System(s):
Method Details:
Experimental Method:
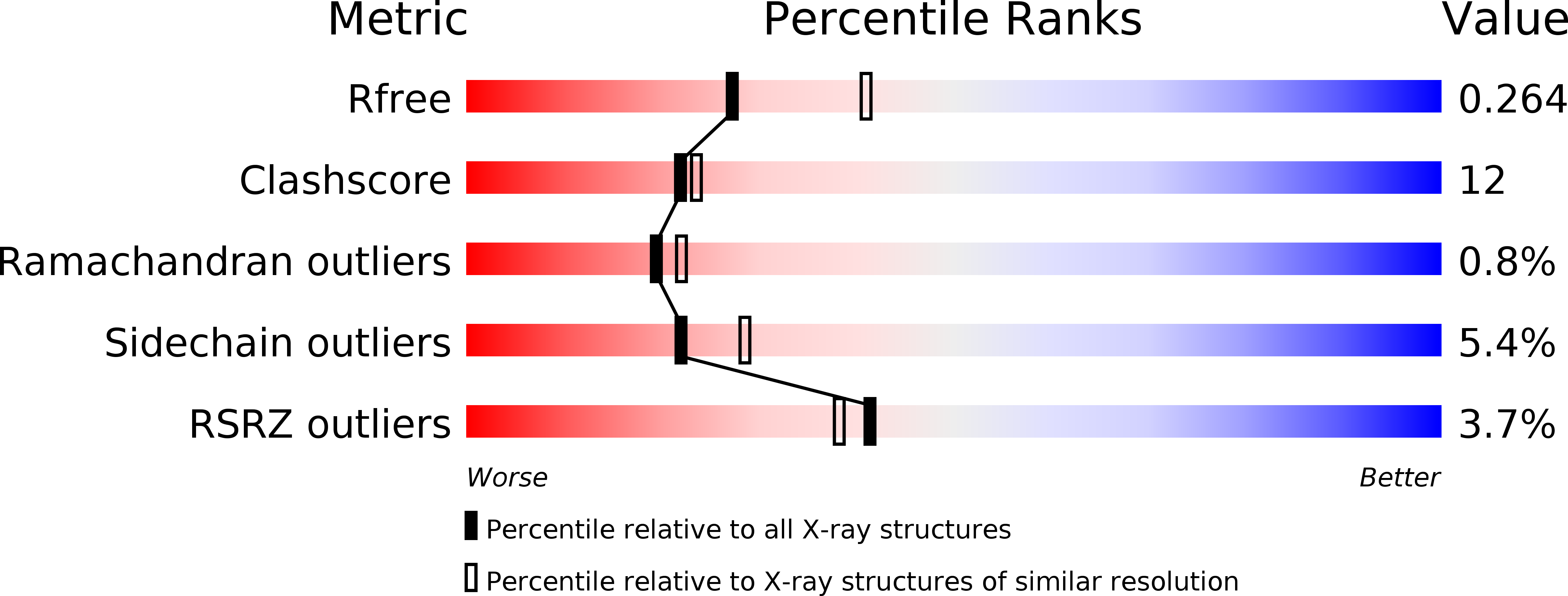
Resolution:
2.45 Å
R-Value Free:
0.26
R-Value Work:
0.24
R-Value Observed:
0.24
Space Group:
P 1 21 1