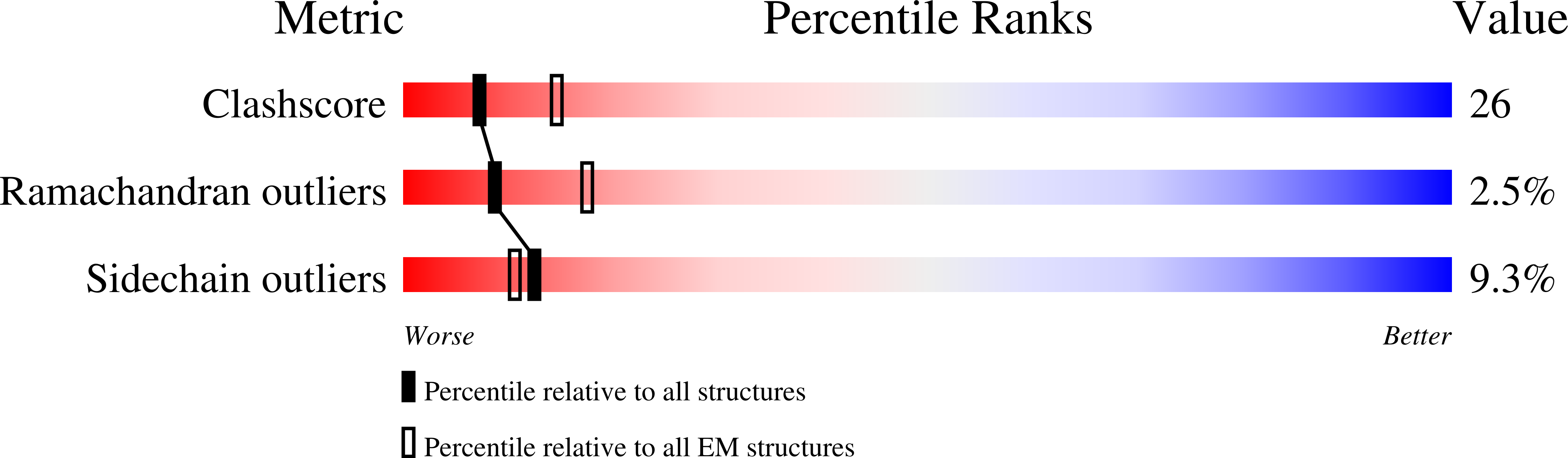
Deposition Date
2003-10-08
Release Date
2004-01-07
Last Version Date
2024-10-09
Entry Detail
PDB ID:
1UPN
Keywords:
Title:
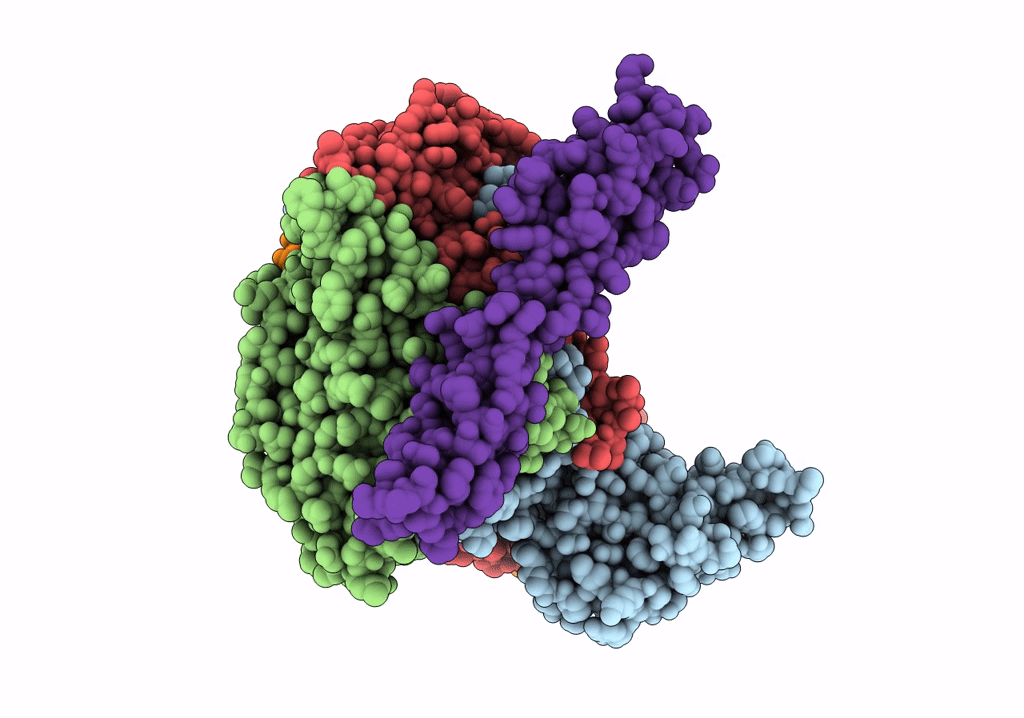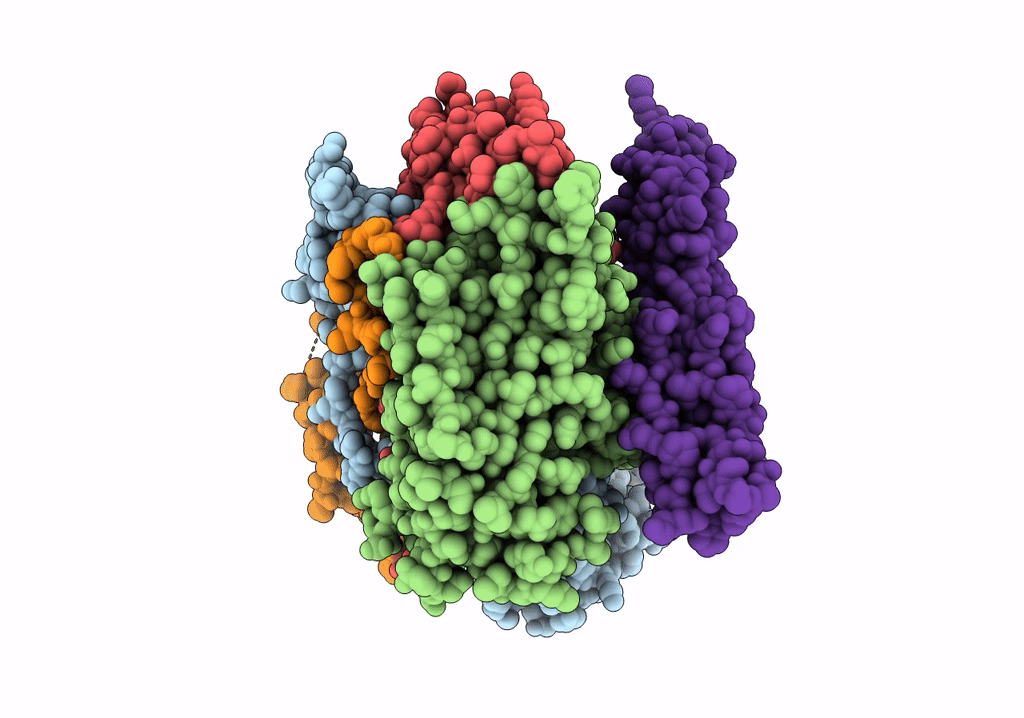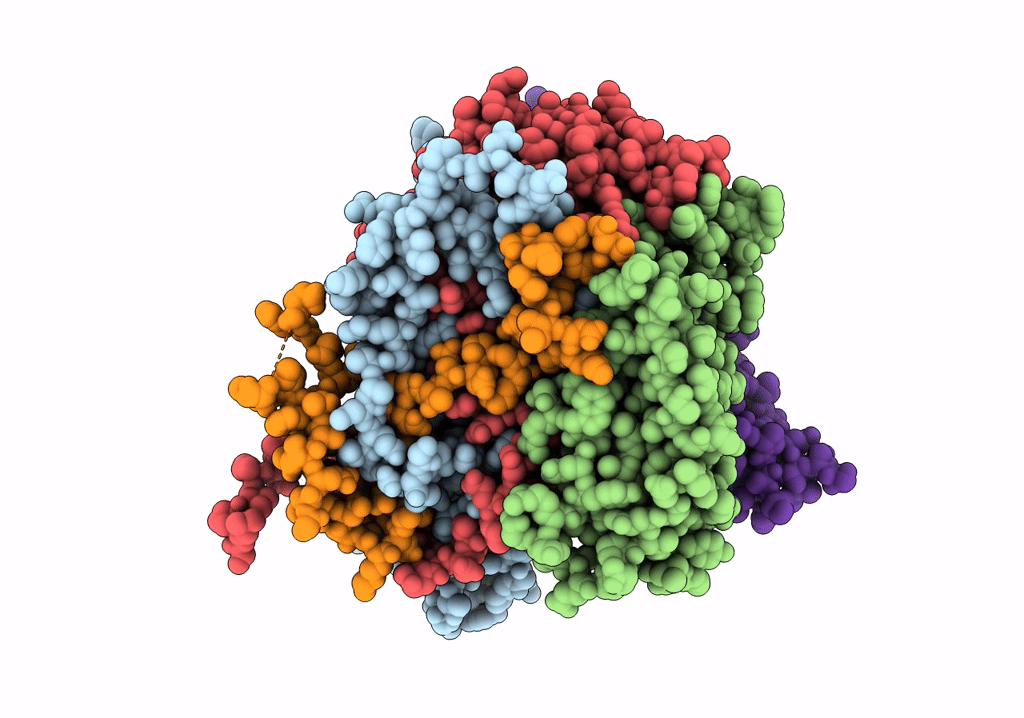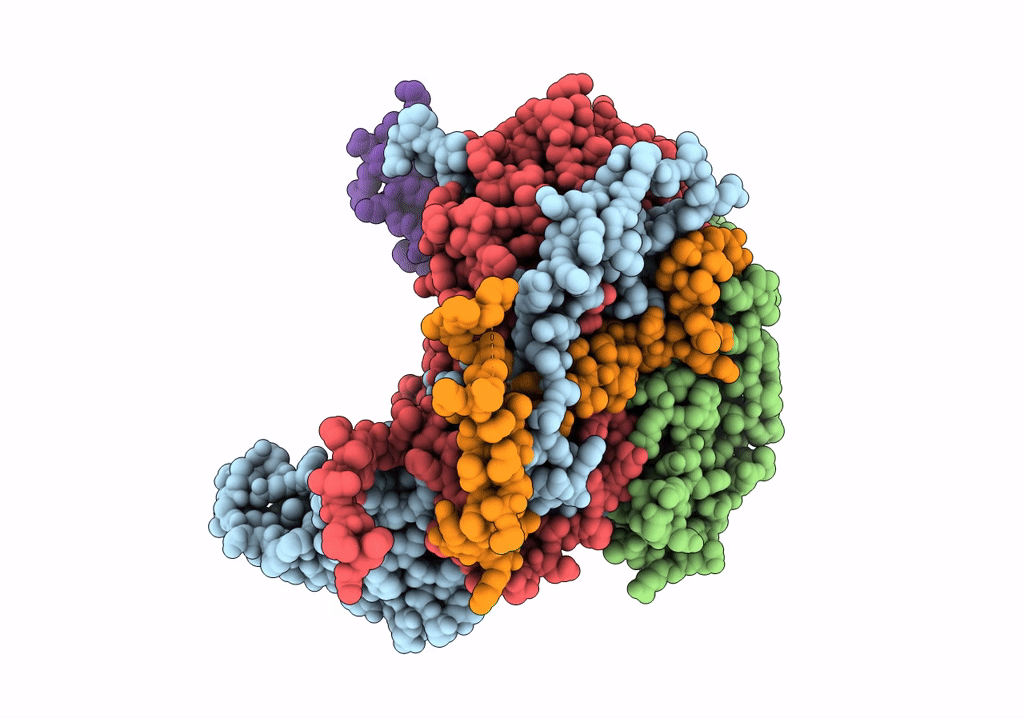
COMPLEX OF ECHOVIRUS TYPE 12 WITH DOMAINS 3 AND 4 OF ITS RECEPTOR DECAY ACCELERATING FACTOR (CD55) BY CRYO ELECTRON MICROSCOPY AT 16 A
Biological Source:
Source Organism(s):
HOMO SAPIENS (Taxon ID: 9606)
HUMAN ECHOVIRUS 11 (Taxon ID: 12078)
HUMAN ECHOVIRUS 11 (Taxon ID: 12078)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
16.00 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE