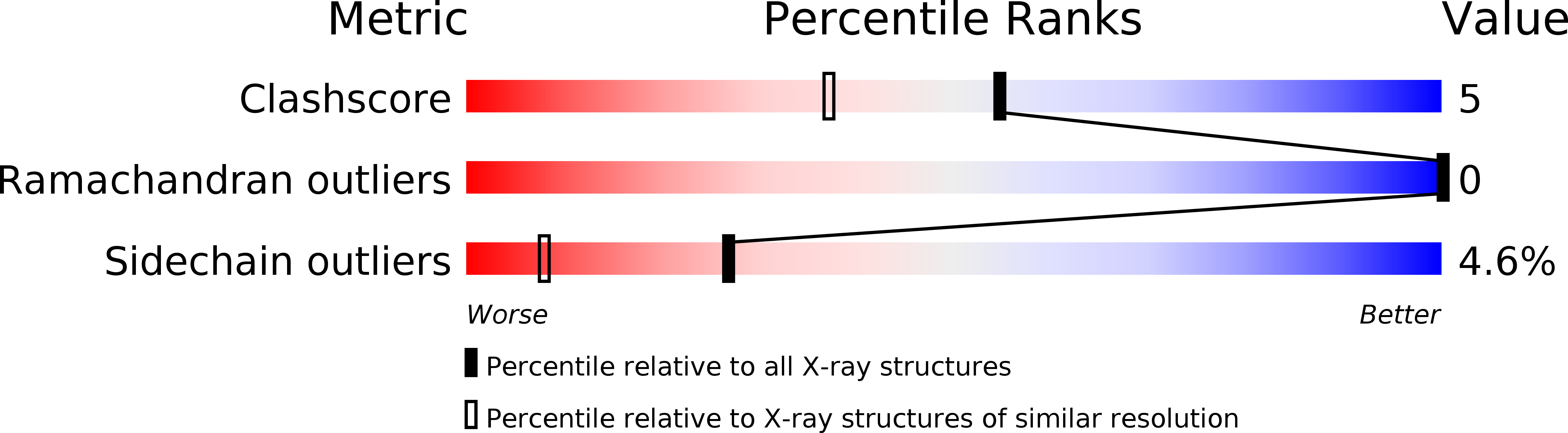
Deposition Date
1995-10-30
Release Date
1996-03-08
Last Version Date
2024-02-14
Entry Detail
PDB ID:
1UDH
Keywords:
Title:
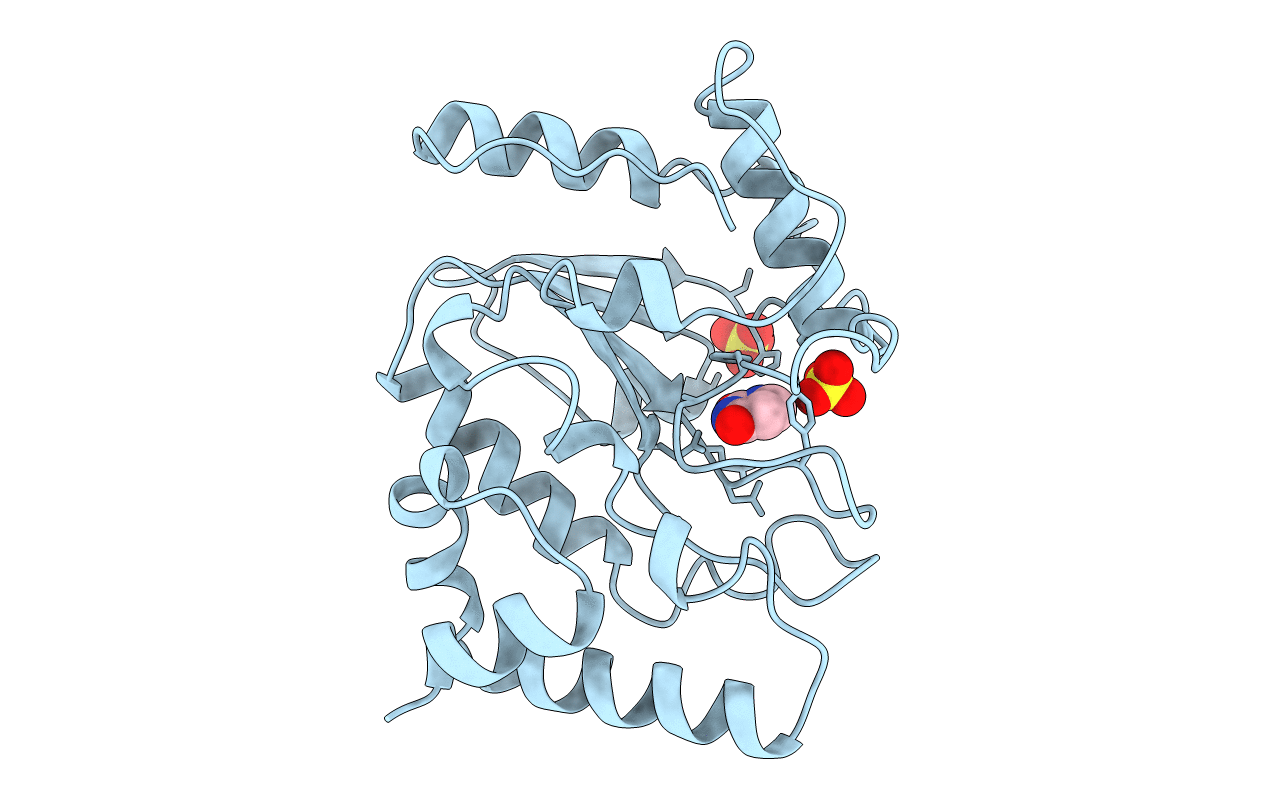
THE STRUCTURAL BASIS OF SPECIFIC BASE EXCISION REPAIR BY URACIL-DNA GLYCOSYLASE
Biological Source:
Source Organism(s):
Herpes simplex virus (type 1 / strain 17) (Taxon ID: 10299)
Method Details:
Experimental Method:
Resolution:
1.75 Å
R-Value Free:
0.23
R-Value Work:
0.19
R-Value Observed:
0.19
Space Group:
P 31