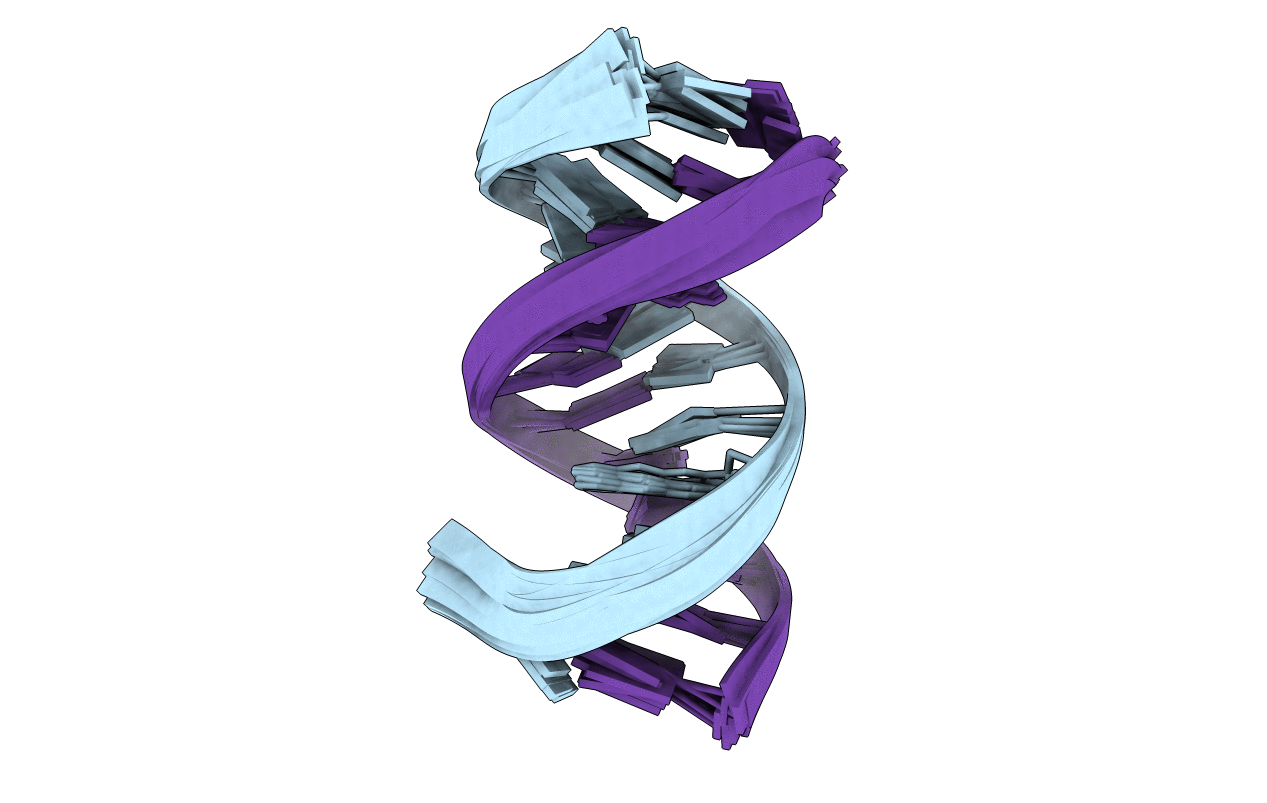
Deposition Date
2004-06-25
Release Date
2004-10-05
Last Version Date
2024-05-22
Entry Detail
PDB ID:
1TUQ
Keywords:
Title:
NMR Structure Analysis of the B-DNA Dodecamer CTCtCACGTGGAG with a tricyclic cytosin base analogue
Biological Source:
Source Organism:
Method Details:
Experimental Method:
Conformers Calculated:
100
Conformers Submitted:
10
Selection Criteria:
lowest CYANA target function


