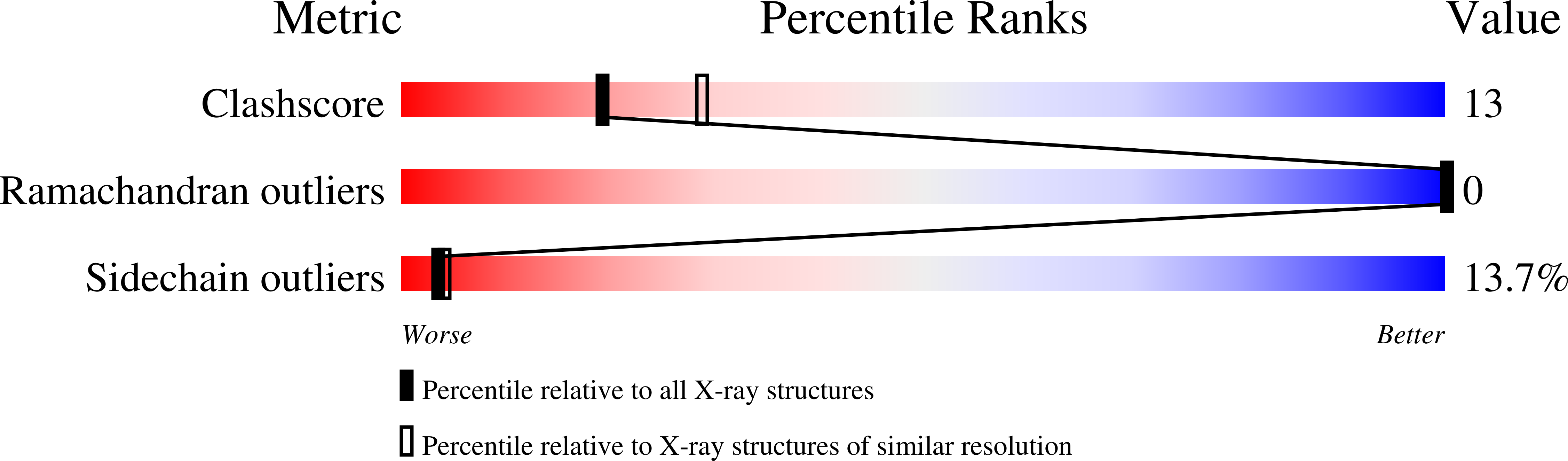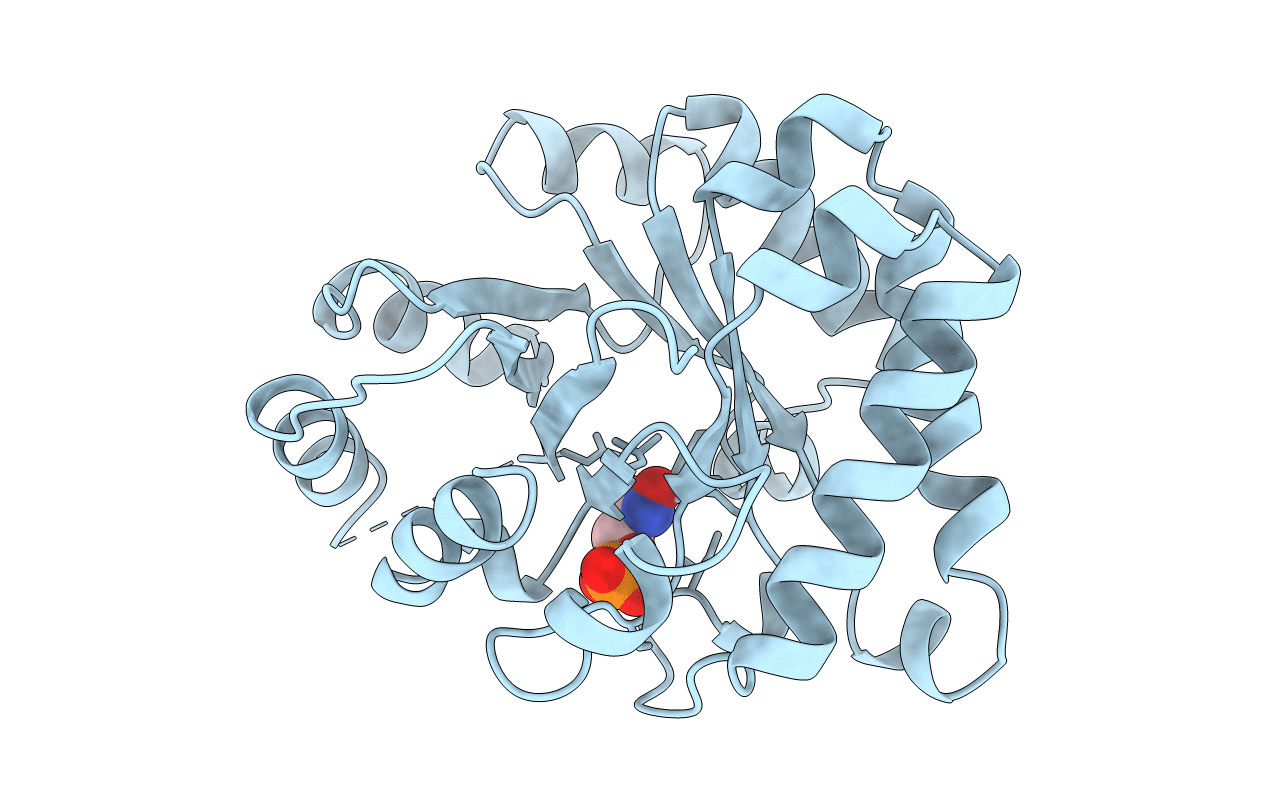
Deposition Date
1995-04-20
Release Date
1995-09-15
Last Version Date
2024-02-14
Entry Detail
PDB ID:
1TTJ
Keywords:
Title:
THREE NEW CRYSTAL STRUCTURES OF POINT MUTATION VARIANTS OF MONOTIM: CONFORMATIONAL FLEXIBILITY OF LOOP-1,LOOP-4 AND LOOP-8
Biological Source:
Source Organism(s):
Trypanosoma brucei brucei (Taxon ID: 5702)
Method Details: