Deposition Date
1994-02-28
Release Date
1994-05-31
Last Version Date
2024-02-14
Entry Detail
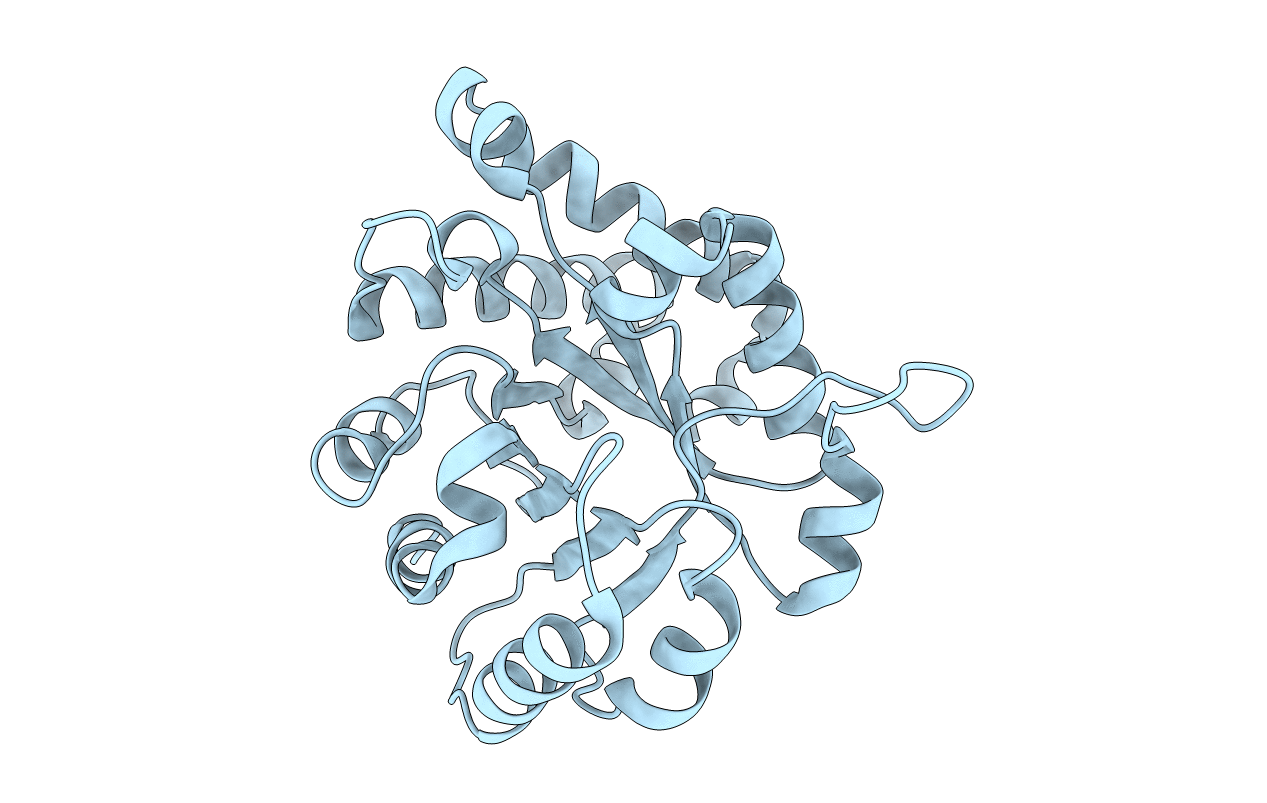
PDB ID:
1TPE
Keywords:
Title:
COMPARISON OF THE STRUCTURES AND THE CRYSTAL CONTACTS OF TRYPANOSOMAL TRIOSEPHOSPHATE ISOMERASE IN FOUR DIFFERENT CRYSTAL FORMS
Biological Source:
Source Organism(s):
Trypanosoma brucei brucei (Taxon ID: 5702)
Method Details:
Experimental Method:
Resolution:
2.10 Å
R-Value Work:
0.15
R-Value Observed:
0.15
Space Group:
C 1 2 1


