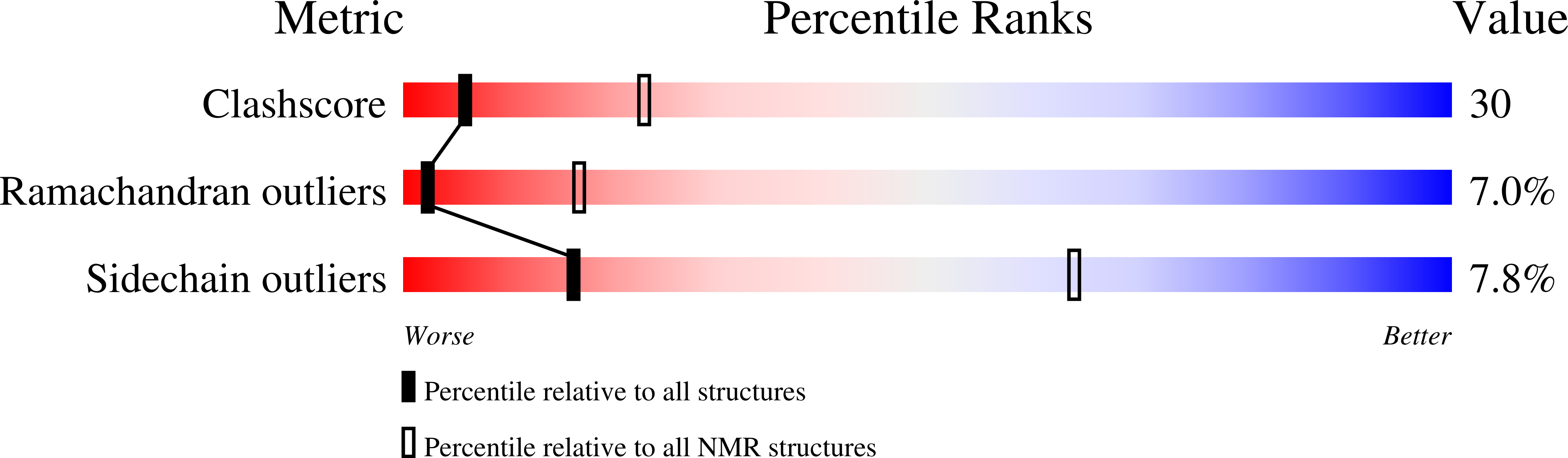
Deposition Date
2004-03-24
Release Date
2004-08-31
Last Version Date
2024-11-20
Entry Detail
PDB ID:
1SSE
Keywords:
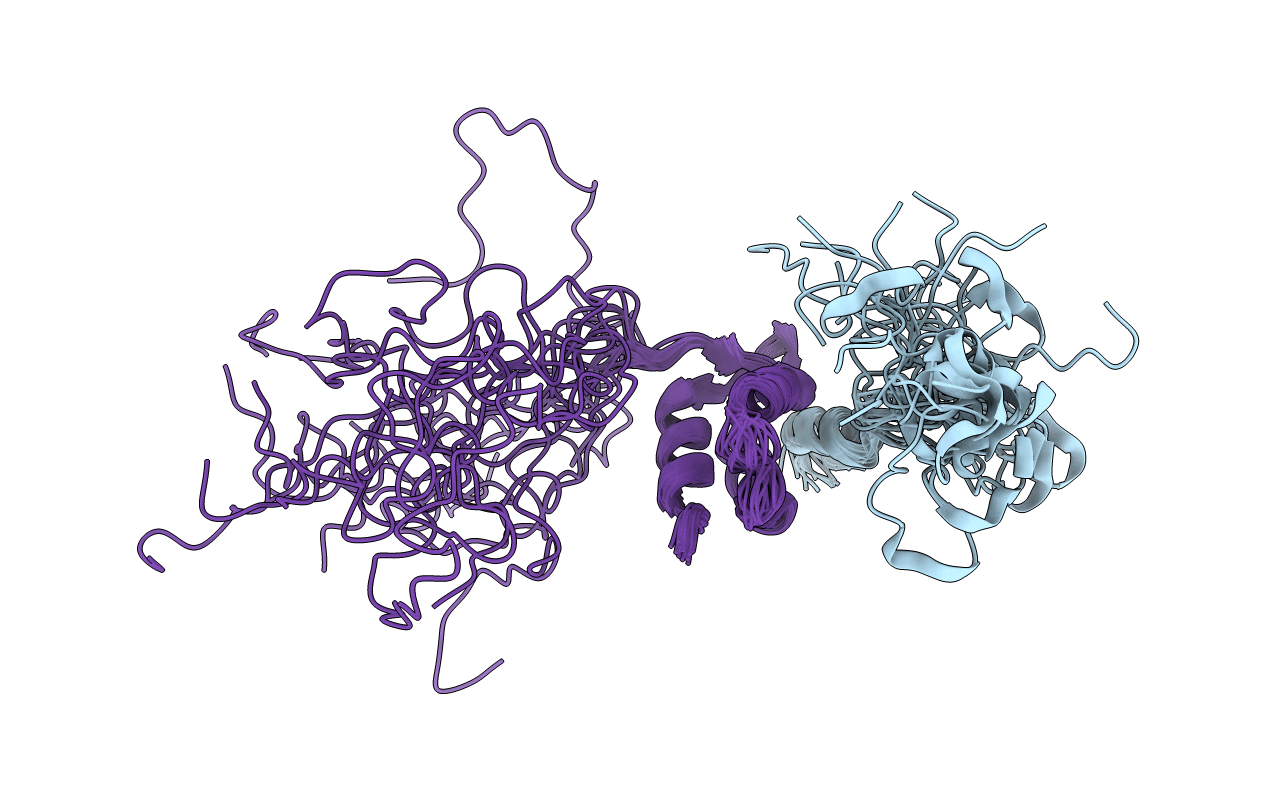
Title:
Solution structure of the oxidized form of the Yap1 redox domain
Biological Source:
Source Organism(s):
Saccharomyces cerevisiae (Taxon ID: 4932)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
100
Conformers Submitted:
20
Selection Criteria:
Stuctures with the lowest energy and no NOE or dihedral violations > 0.5 A and 5 degrees, respectively.