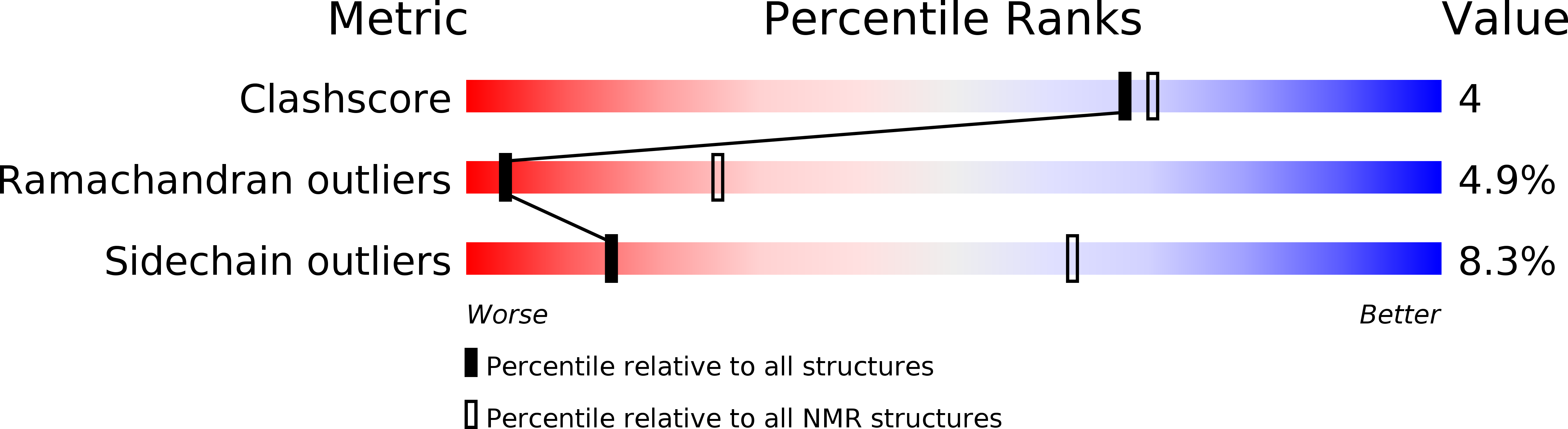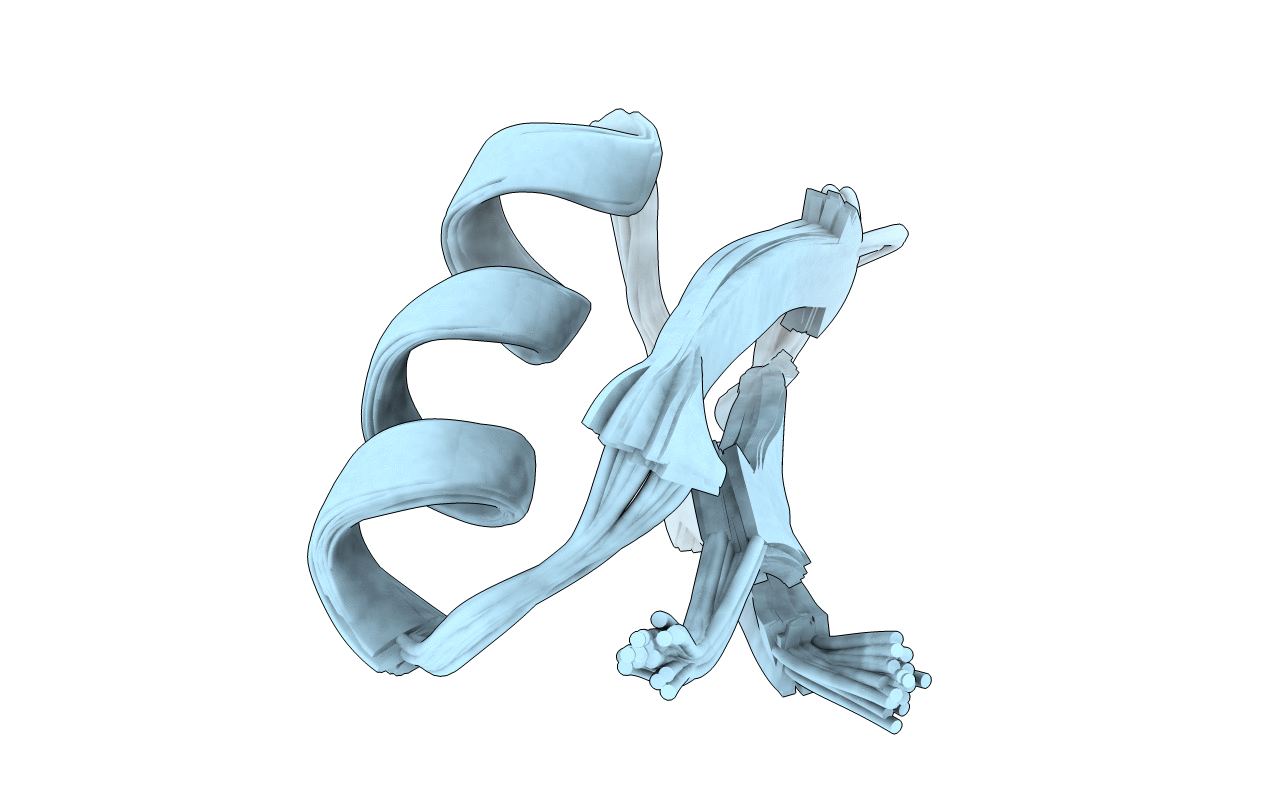
Deposition Date
1996-04-01
Release Date
1997-01-27
Last Version Date
2024-11-13
Entry Detail
PDB ID:
1SCO
Keywords:
Title:
SCORPION TOXIN (OSK1 TOXIN) WITH HIGH AFFINITY FOR SMALL CONDUCTANCE CA(2+)-ACTIVATED K+ CHANNEL IN NEUROBLASTOMA-X-GLUOMA NG 108-15 HYBRID CELLS, NMR, 30 STRUCTURES
Biological Source:
Source Organism(s):
Orthochirus scrobiculosus (Taxon ID: 6892)
Method Details:
Experimental Method:
Conformers Submitted:
30