Deposition Date
2004-02-06
Release Date
2004-09-07
Last Version Date
2024-02-14
Entry Detail
PDB ID:
1SA3
Keywords:
Title:
An asymmetric complex of restriction endonuclease MspI on its palindromic DNA recognition site
Biological Source:
Source Organism(s):
Moraxella sp. (Taxon ID: 479)
Expression System(s):
Method Details:
Experimental Method:
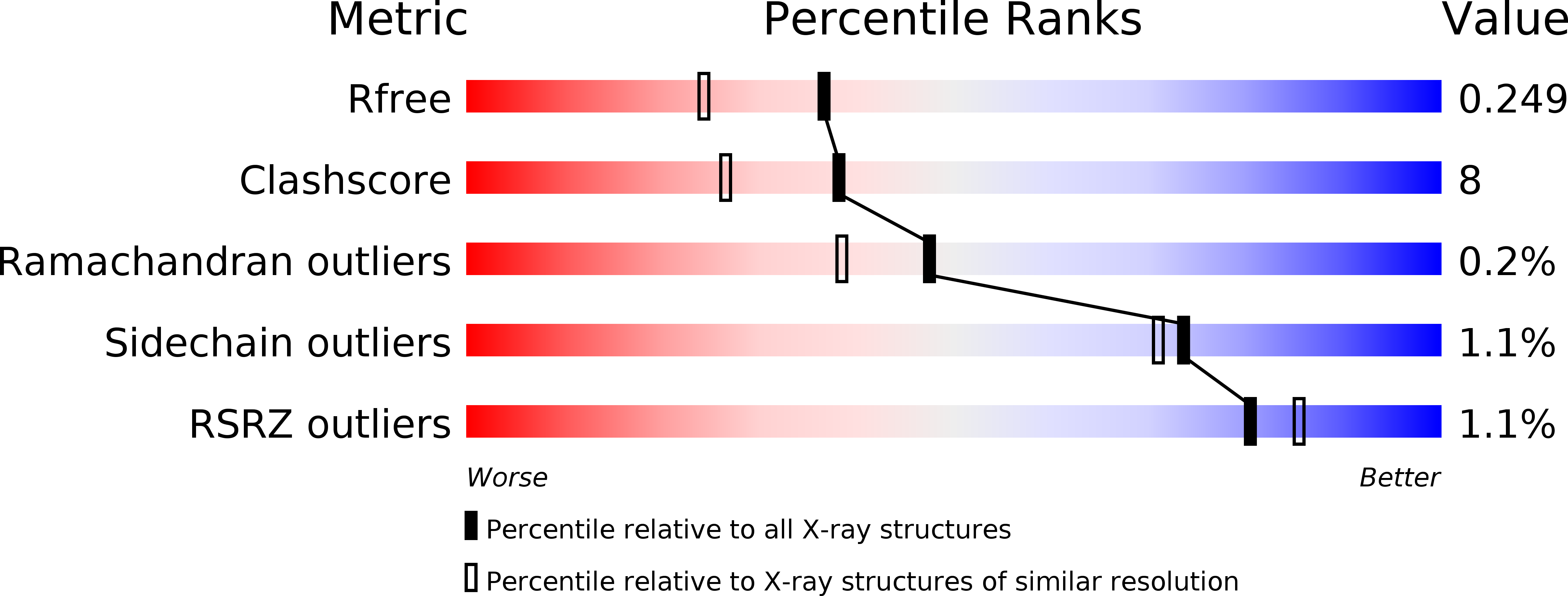
Resolution:
1.95 Å
R-Value Free:
0.25
R-Value Work:
0.22
R-Value Observed:
0.22
Space Group:
P 1 21 1