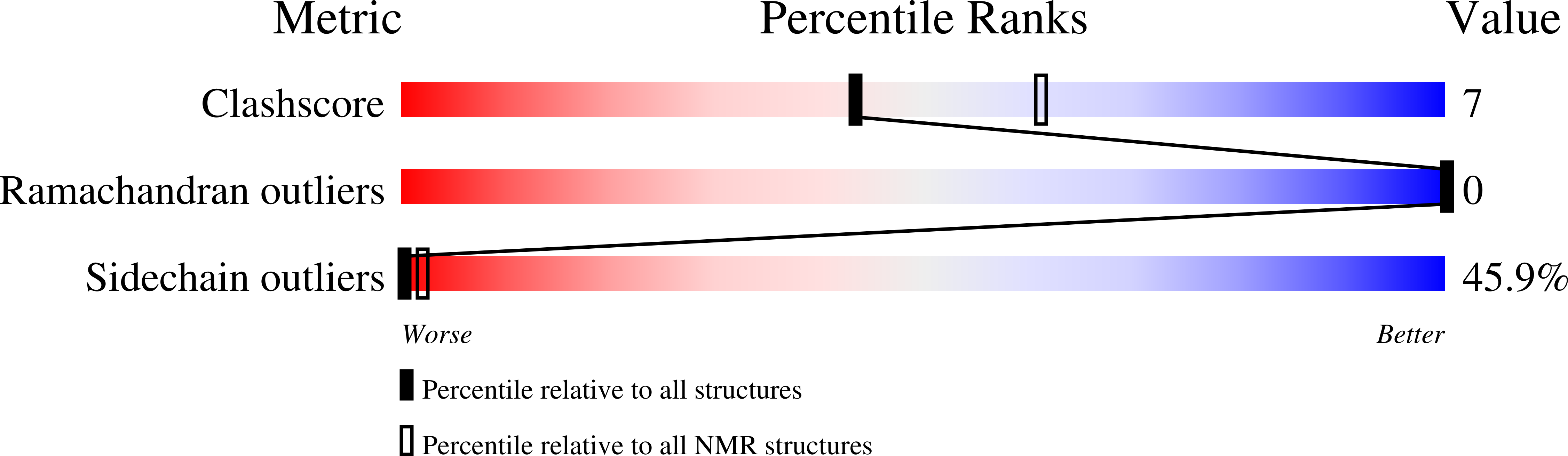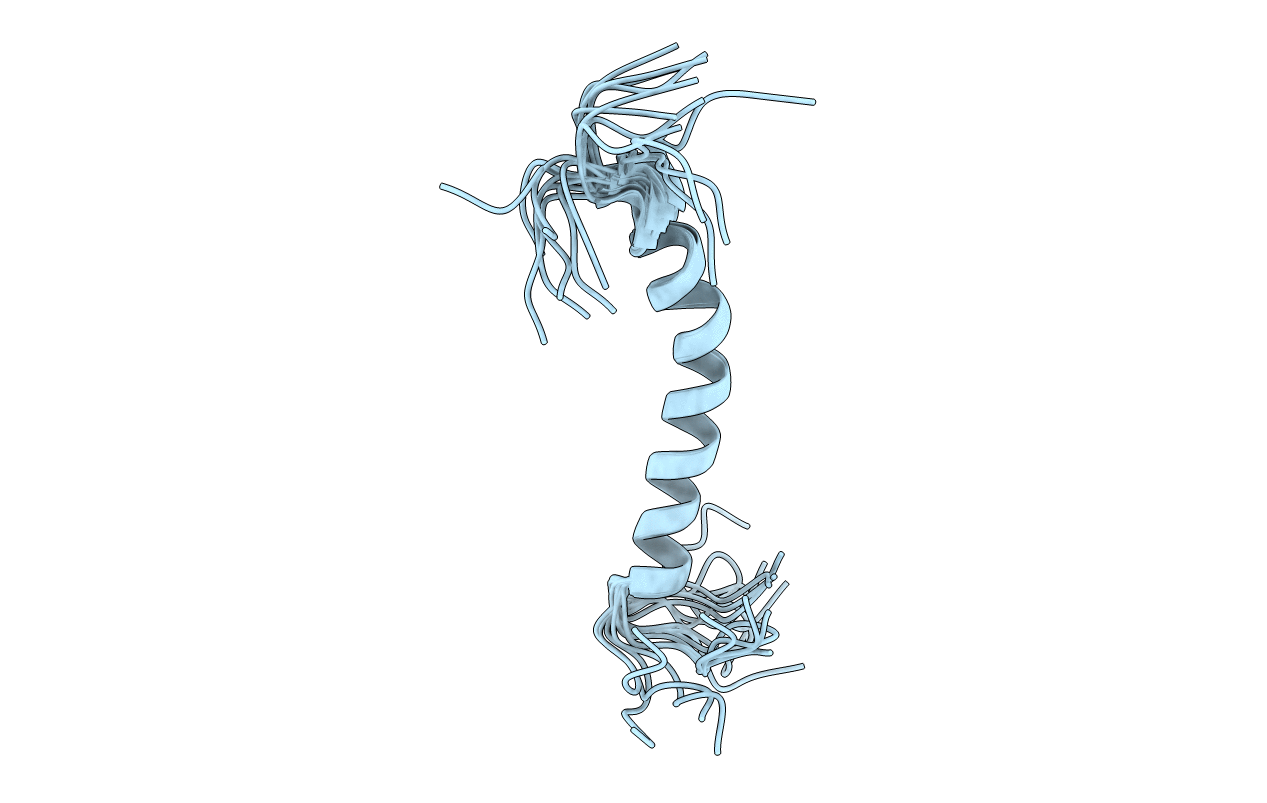
Deposition Date
2003-11-21
Release Date
2004-03-23
Last Version Date
2024-05-22
Entry Detail
PDB ID:
1RKL
Keywords:
Title:
NMR structure of yeast oligosaccharyltransferase subunit Ost4p
Method Details:
Experimental Method:
Conformers Calculated:
500
Conformers Submitted:
20
Selection Criteria:
structures with the lowest energy