Deposition Date
1996-12-06
Release Date
1997-06-16
Last Version Date
2024-10-30
Entry Detail
PDB ID:
1RCX
Keywords:
Title:
NON-ACTIVATED SPINACH RUBISCO IN COMPLEX WITH ITS SUBSTRATE RIBULOSE-1,5-BISPHOSPHATE
Biological Source:
Source Organism(s):
Spinacia oleracea (Taxon ID: 3562)
Method Details:
Experimental Method:
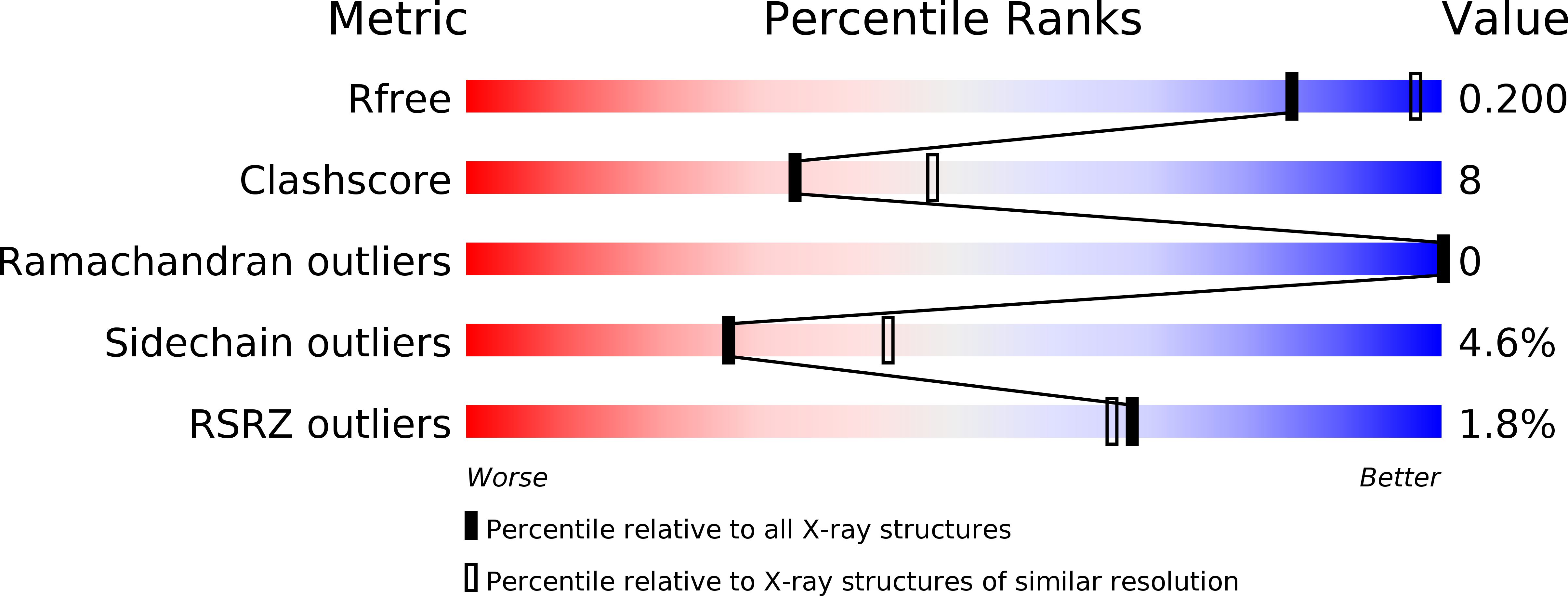
Resolution:
2.40 Å
R-Value Free:
0.23
R-Value Work:
0.22
R-Value Observed:
0.22
Space Group:
P 21 21 2