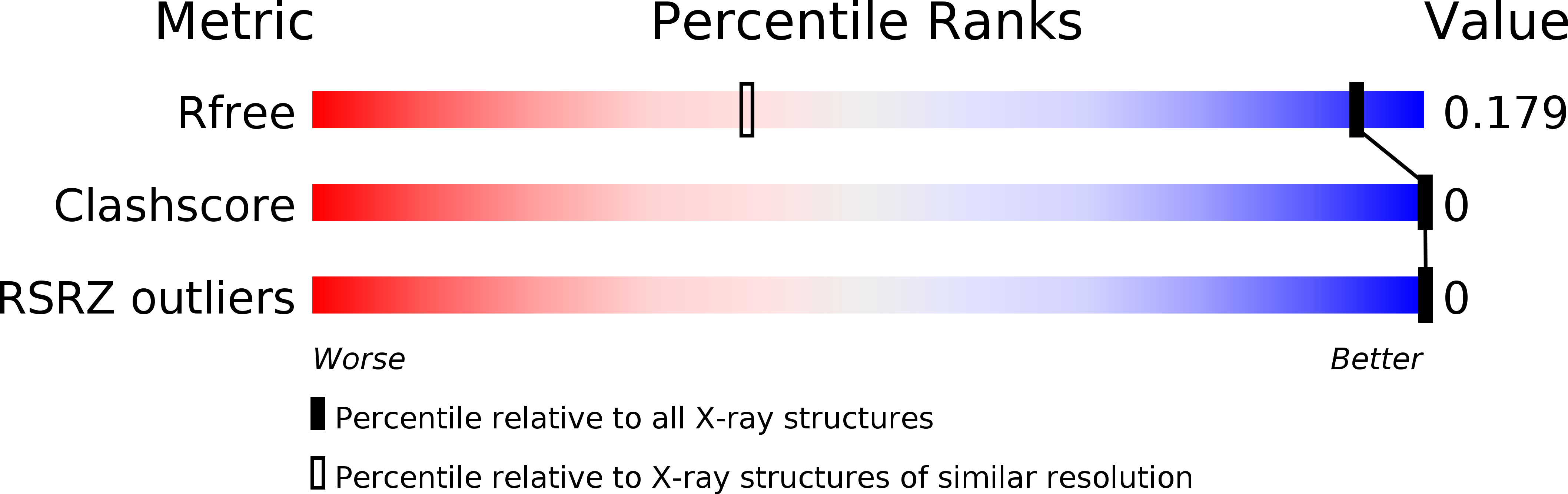
Deposition Date
2003-10-01
Release Date
2003-10-21
Last Version Date
2024-02-14
Entry Detail
PDB ID:
1R3G
Keywords:
Title:
1.16A X-ray structure of the synthetic DNA fragment with the incorporated 2'-O-[(2-Guanidinium)ethyl]-5-methyluridine residues
Method Details:
Experimental Method:
Resolution:
1.16 Å
R-Value Free:
0.17
R-Value Work:
0.13
R-Value Observed:
0.13
Space Group:
P 21 21 21