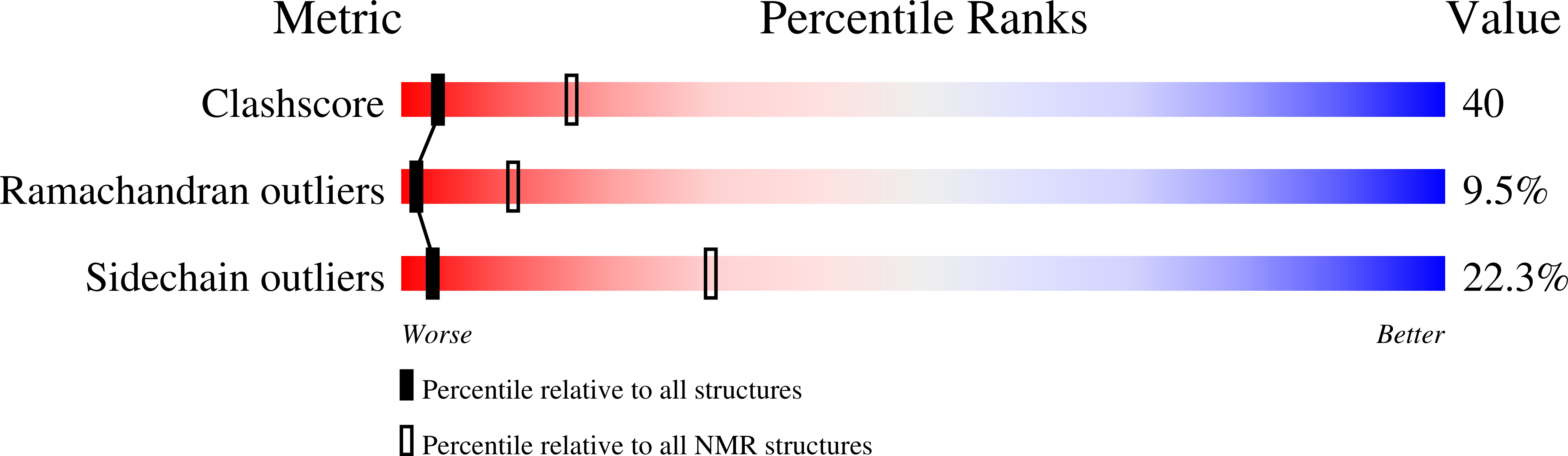Deposition Date
1999-07-08
Release Date
1999-12-23
Last Version Date
2023-12-27
Entry Detail
PDB ID:
1QU6
Keywords:
Title:
STRUCTURE OF THE DOUBLE-STRANDED RNA-BINDING DOMAIN OF THE PROTEIN KINASE PKR REVEALS THE MOLECULAR BASIS OF ITS DSRNA-MEDIATED ACTIVATION
Biological Source:
Source Organism:
Homo sapiens (Taxon ID: 9606)
Host Organism:
Method Details: