Deposition Date
1996-01-04
Release Date
1996-06-10
Last Version Date
2024-02-14
Entry Detail
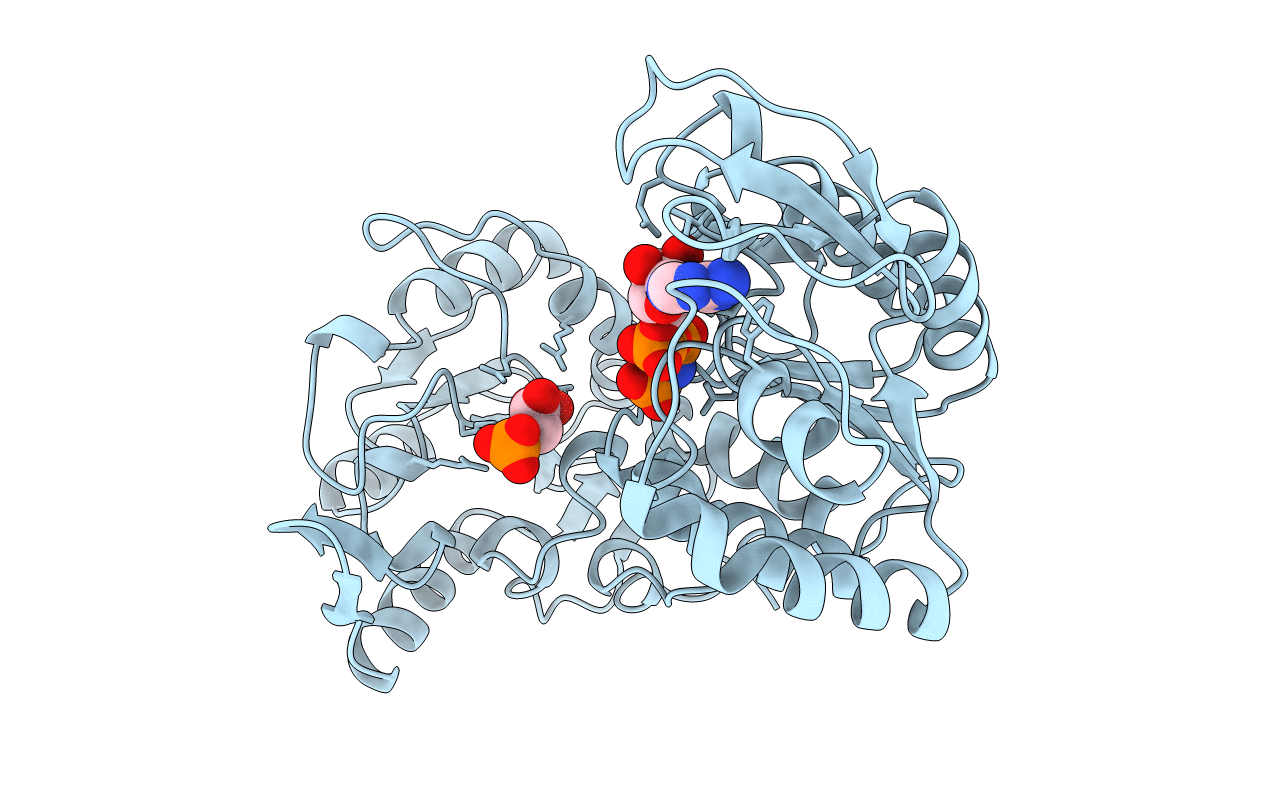
PDB ID:
1QPG
Keywords:
Title:
3-PHOSPHOGLYCERATE KINASE, MUTATION R65Q
Biological Source:
Source Organism:
Saccharomyces cerevisiae (Taxon ID: 4932)
Host Organism:
Method Details:
Experimental Method:
Resolution:
2.40 Å
R-Value Work:
0.18
R-Value Observed:
0.18
Space Group:
C 1 2 1


