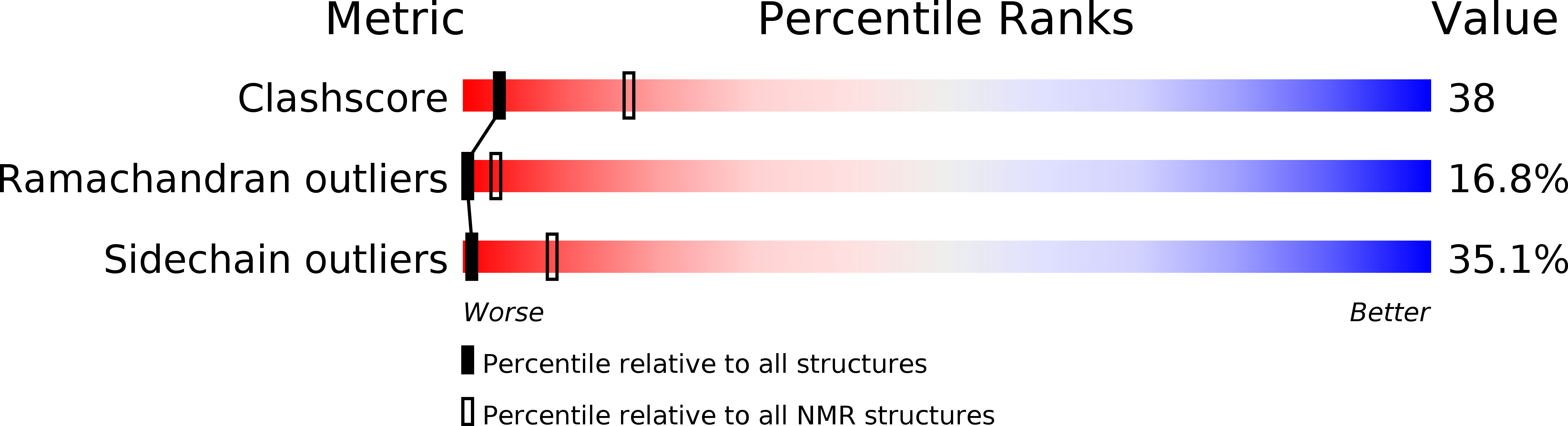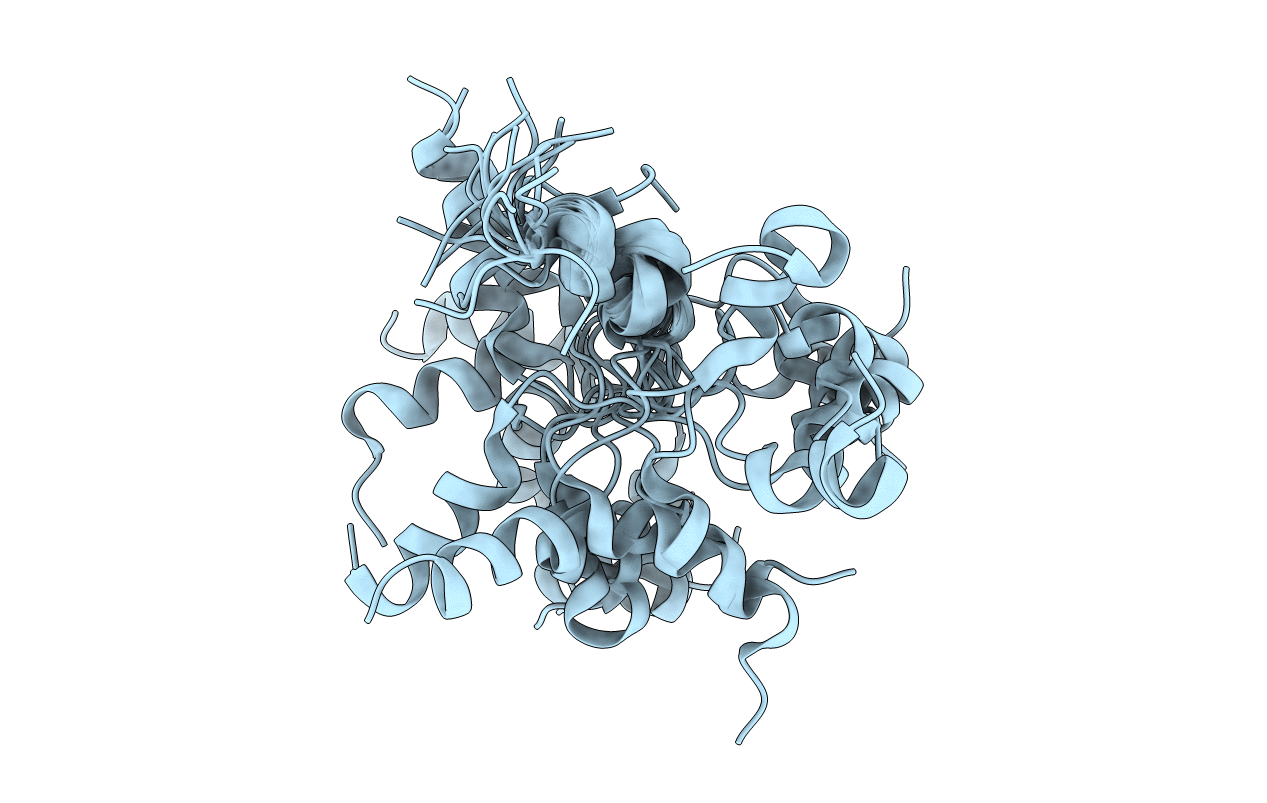
Deposition Date
1999-09-09
Release Date
1999-12-14
Last Version Date
2024-05-15
Entry Detail
PDB ID:
1QLO
Keywords:
Title:
Structure of the active domain of the herpes simplex virus protein ICP47 in water/sodium dodecyl sulfate solution determined by nuclear magnetic resonance spectroscopy
Biological Source:
Source Organism(s):
HERPES SIMPLEX VIRUS (Taxon ID: 10299)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
40
Conformers Submitted:
19
Selection Criteria:
LEAST RESTRAINT VIOLATION ENERGY