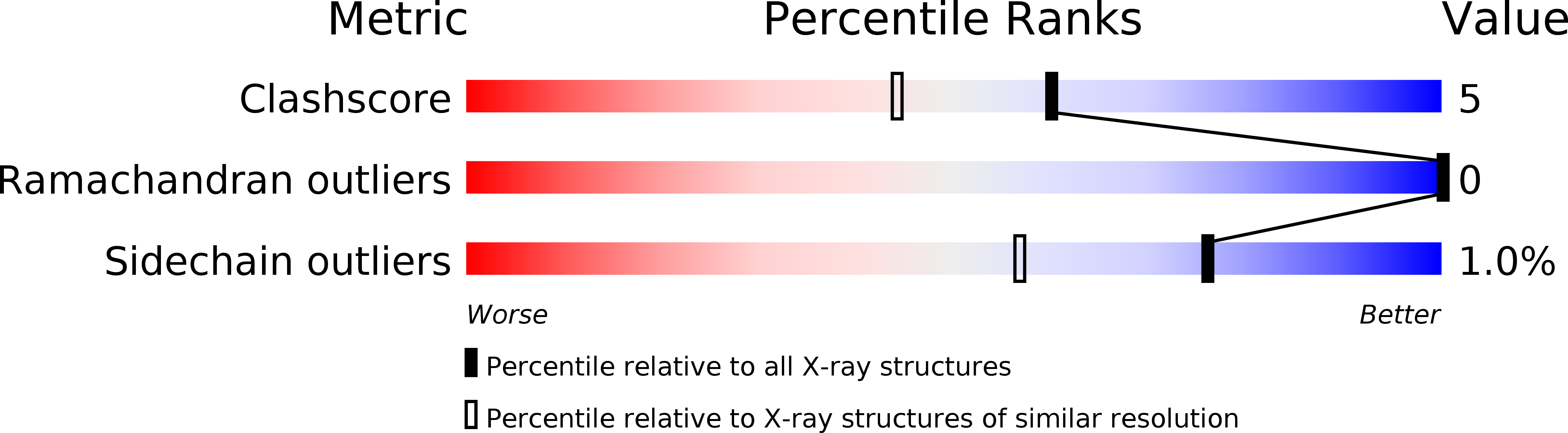
Deposition Date
1999-08-20
Release Date
2000-02-03
Last Version Date
2024-11-06
Entry Detail
PDB ID:
1QL4
Keywords:
Title:
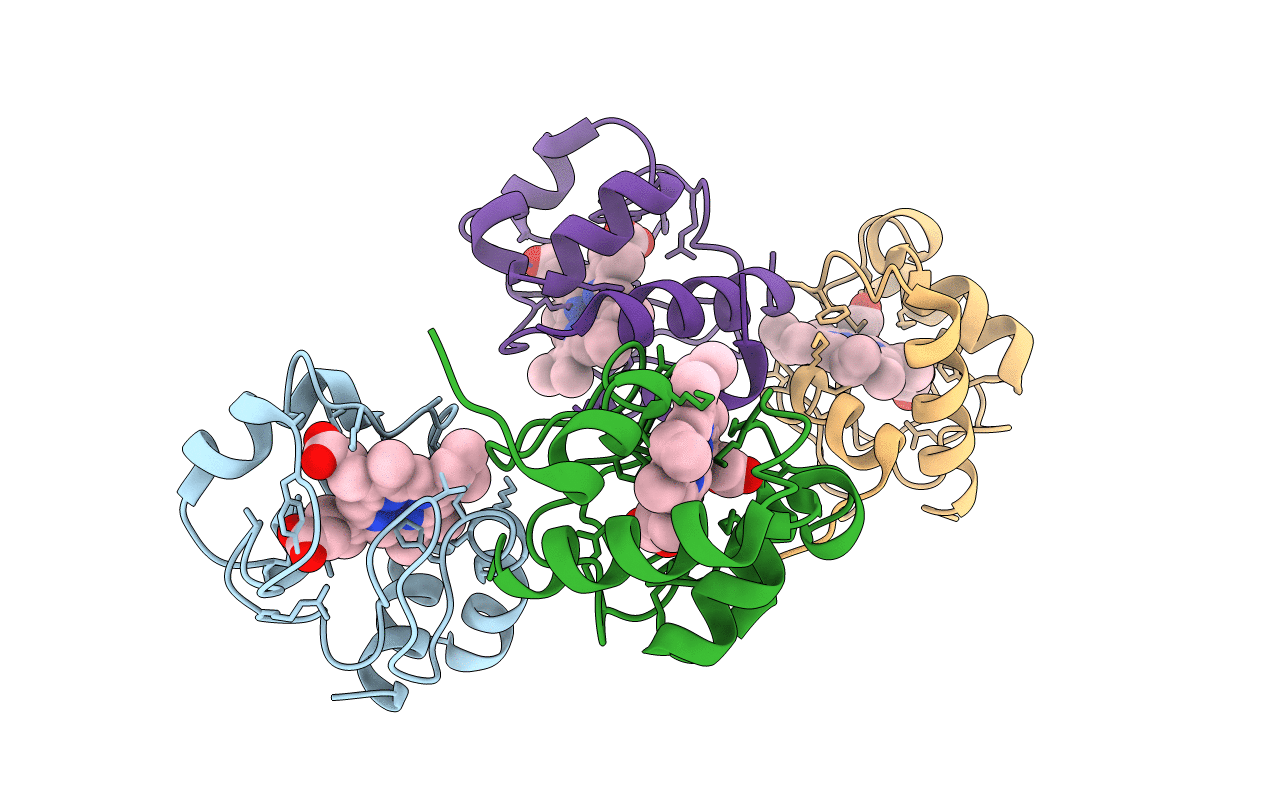
Structure of the soluble domain of cytochrome c552 from Paracoccus denitrificans in the oxidised state
Biological Source:
Source Organism(s):
PARACOCCUS DENITRIFICANS (Taxon ID: 266)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
1.50 Å
R-Value Free:
0.26
R-Value Work:
0.23
R-Value Observed:
0.23
Space Group:
P 21 21 21