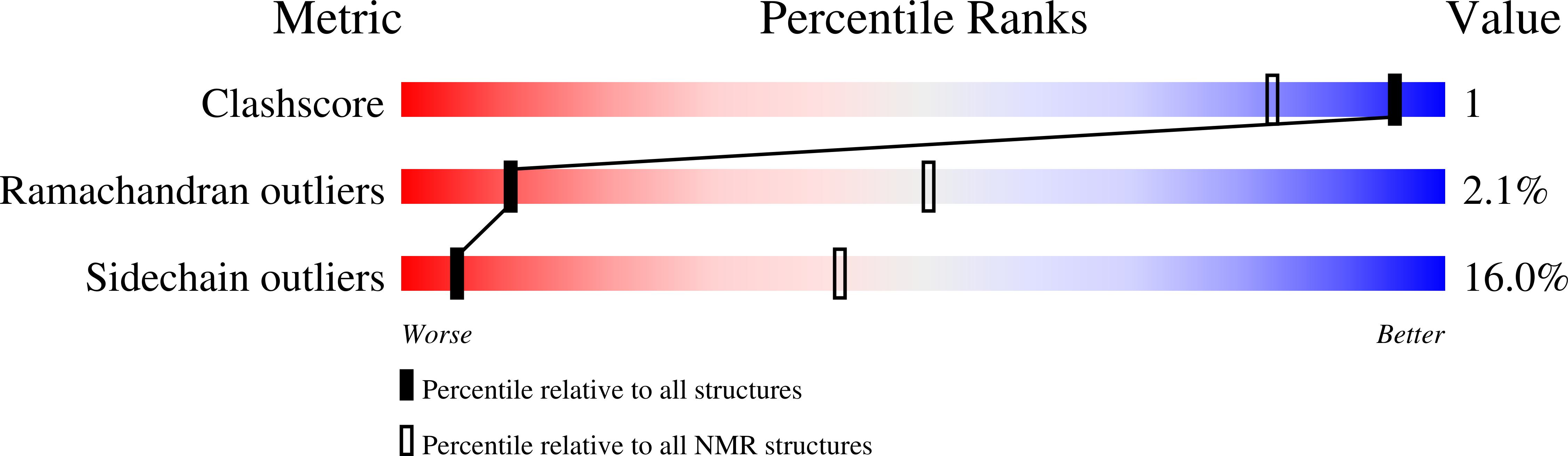
Deposition Date
1999-04-12
Release Date
2000-01-01
Last Version Date
2024-11-13
Entry Detail
PDB ID:
1QFN
Keywords:
Title:
GLUTAREDOXIN-1-RIBONUCLEOTIDE REDUCTASE B1 MIXED DISULFIDE BOND
Biological Source:
Source Organism(s):
Escherichia coli (Taxon ID: 562)
Expression System(s):
Method Details:
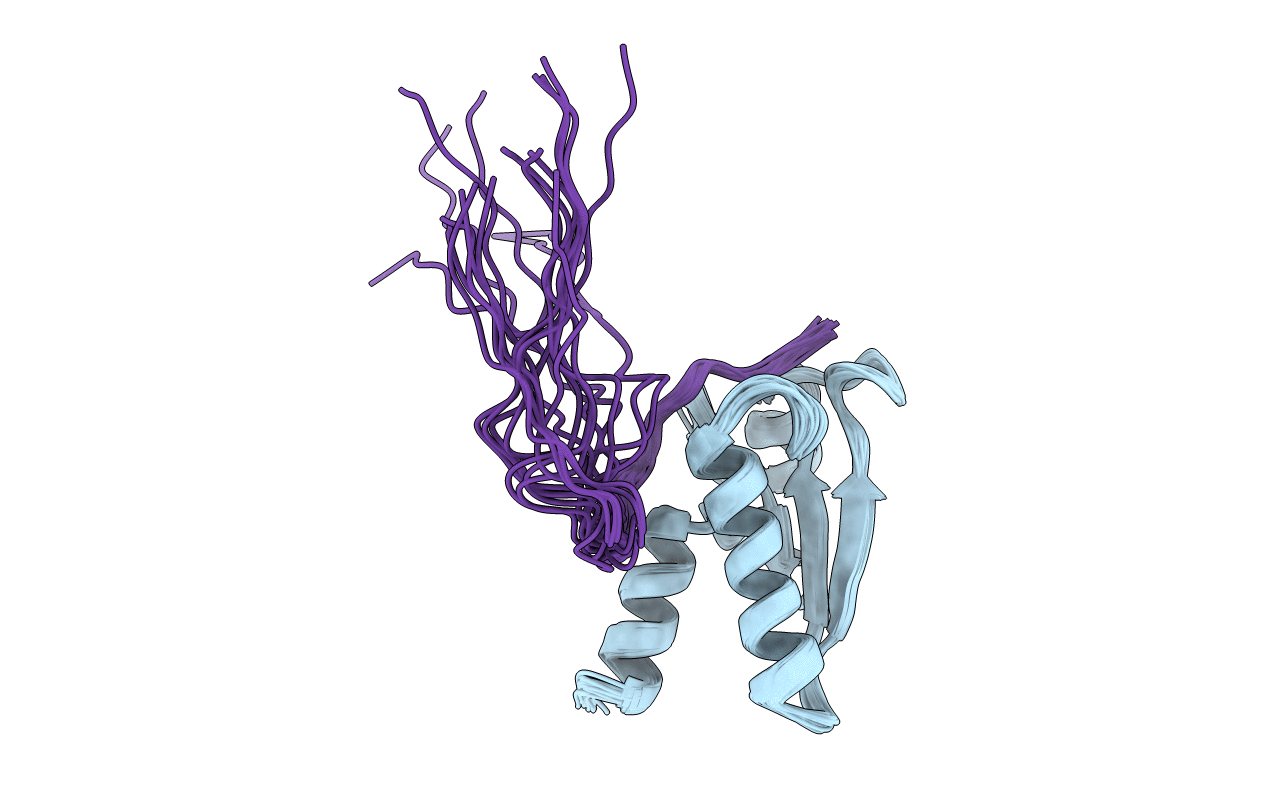
Experimental Method:
Conformers Calculated:
50
Conformers Submitted:
20
Selection Criteria:
LEAST TARGET FUNCTION