Deposition Date
2003-08-22
Release Date
2004-06-01
Last Version Date
2024-11-13
Entry Detail
PDB ID:
1Q94
Keywords:
Title:
Structures of HLA-A*1101 in complex with immunodominant nonamer and decamer HIV-1 epitopes clearly reveal the presence of a middle anchor residue
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Human immunodeficiency virus type 1 group M subtype D (isolate Z2/CDC-Z34) (HIV-1) (Taxon ID: 11683)
Human immunodeficiency virus type 1 group M subtype D (isolate Z2/CDC-Z34) (HIV-1) (Taxon ID: 11683)
Expression System(s):
Method Details:
Experimental Method:
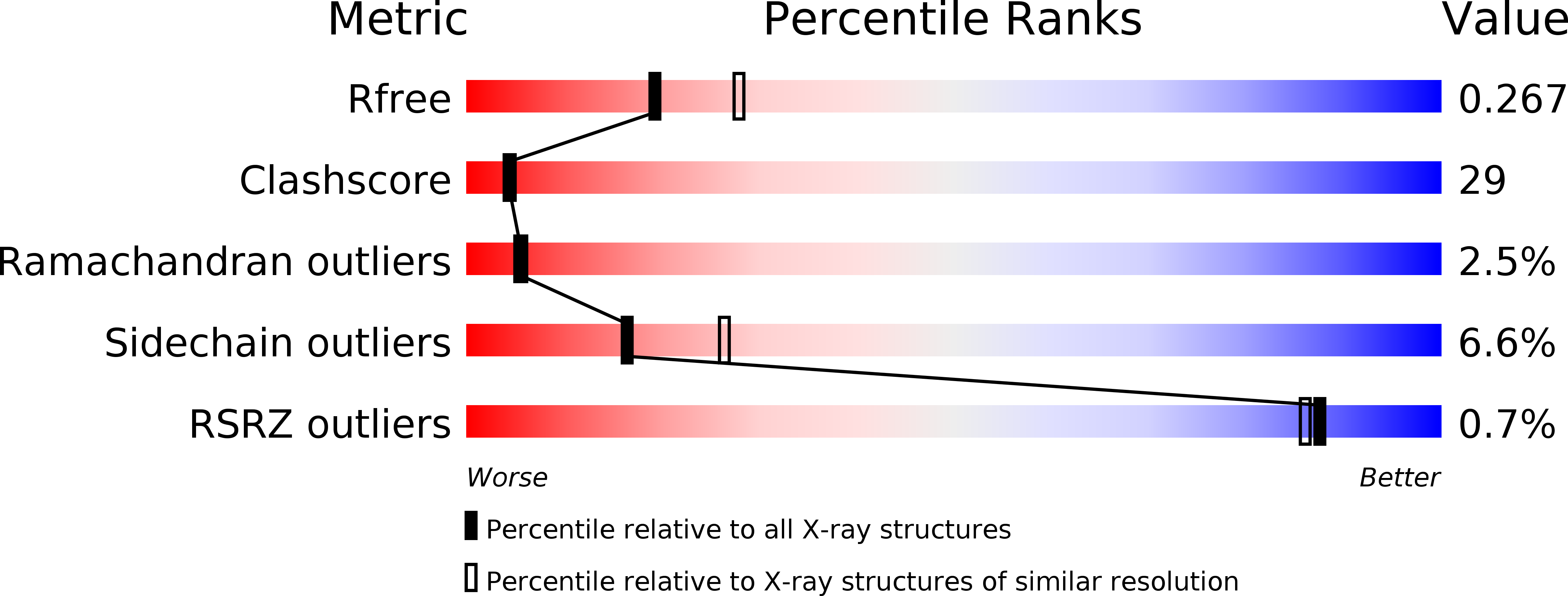
Resolution:
2.40 Å
R-Value Free:
0.27
R-Value Work:
0.23
R-Value Observed:
0.23
Space Group:
P 1