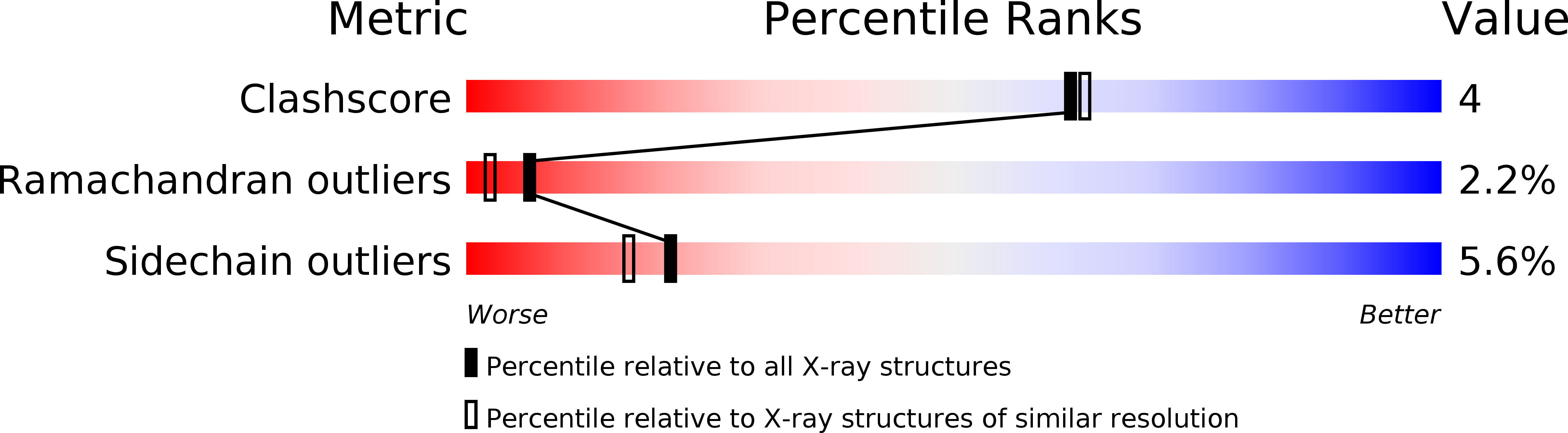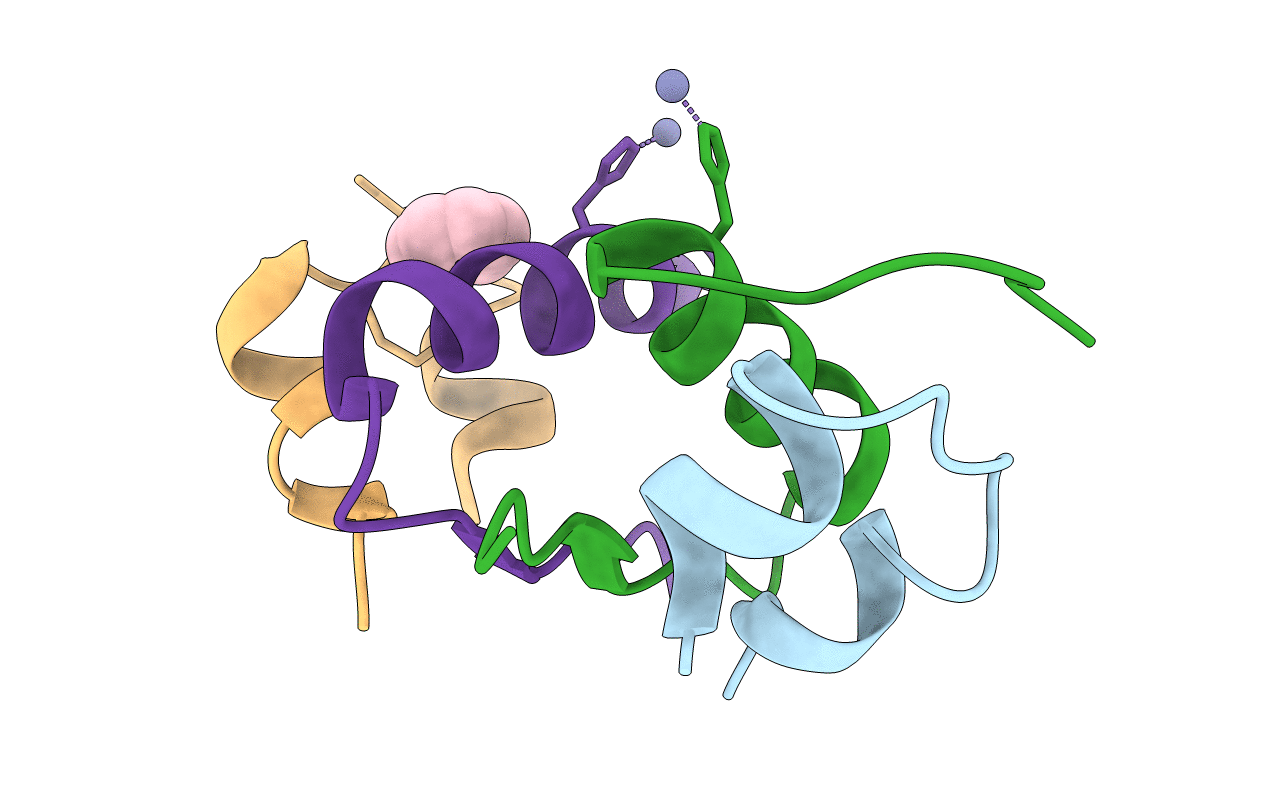
Deposition Date
2003-08-04
Release Date
2003-08-19
Last Version Date
2024-11-20
Entry Detail
PDB ID:
1Q4V
Keywords:
Title:
CRYSTAL STRUCTURE OF ALLO-ILEA2-INSULIN, AN INACTIVE CHIRAL ANALOGUE: IMPLICATIONS FOR THE MECHANISM OF RECEPTOR
Method Details:
Experimental Method:
Resolution:
2.00 Å
R-Value Free:
0.26
R-Value Work:
0.21
Space Group:
H 3