Deposition Date
2003-07-24
Release Date
2003-11-25
Last Version Date
2024-11-20
Entry Detail
PDB ID:
1Q2B
Keywords:
Title:
CELLOBIOHYDROLASE CEL7A WITH DISULPHIDE BRIDGE ADDED ACROSS EXO-LOOP BY MUTATIONS D241C AND D249C
Biological Source:
Source Organism(s):
Hypocrea jecorina (Taxon ID: 51453)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
1.60 Å
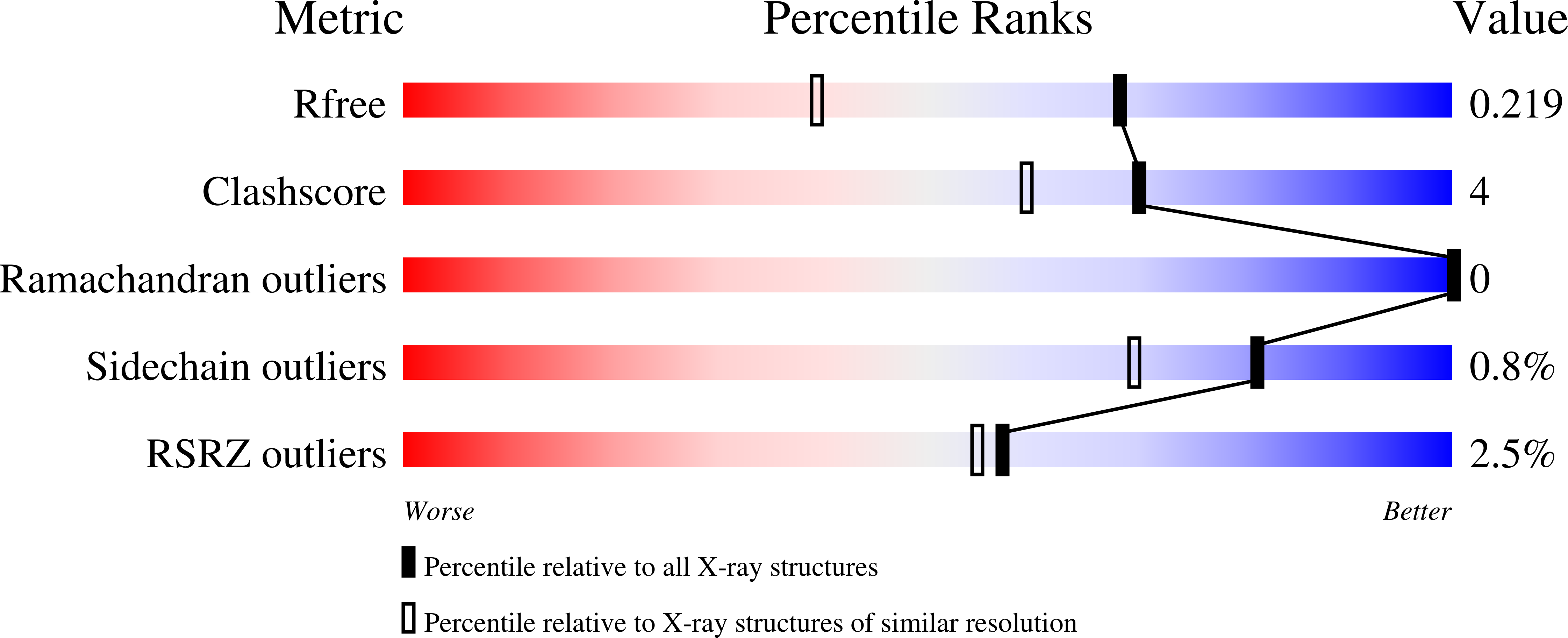
R-Value Free:
0.22
R-Value Work:
0.20
Space Group:
I 2 2 2