Deposition Date
2003-07-09
Release Date
2004-04-20
Last Version Date
2024-05-01
Entry Detail
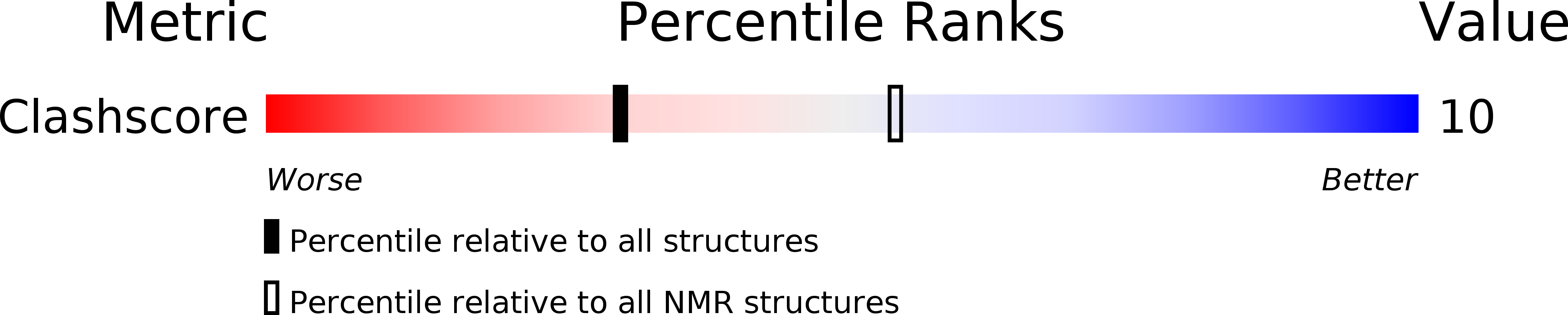
PDB ID:
1PYJ
Keywords:
Title:
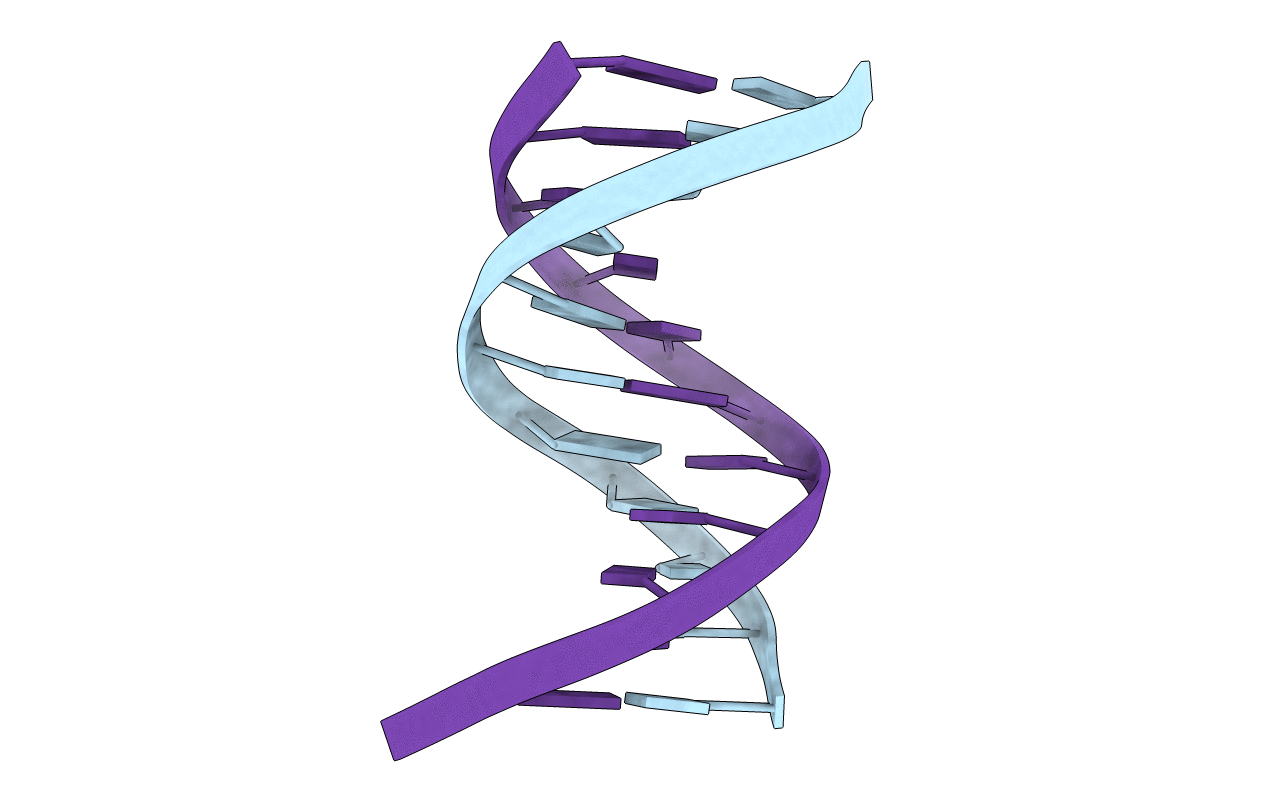
Solution Structure of an O6-[4-oxo-4-(3-pyridyl)butyl]guanine adduct in an 11mer DNA duplex
Method Details:
Experimental Method:
Conformers Submitted:
1