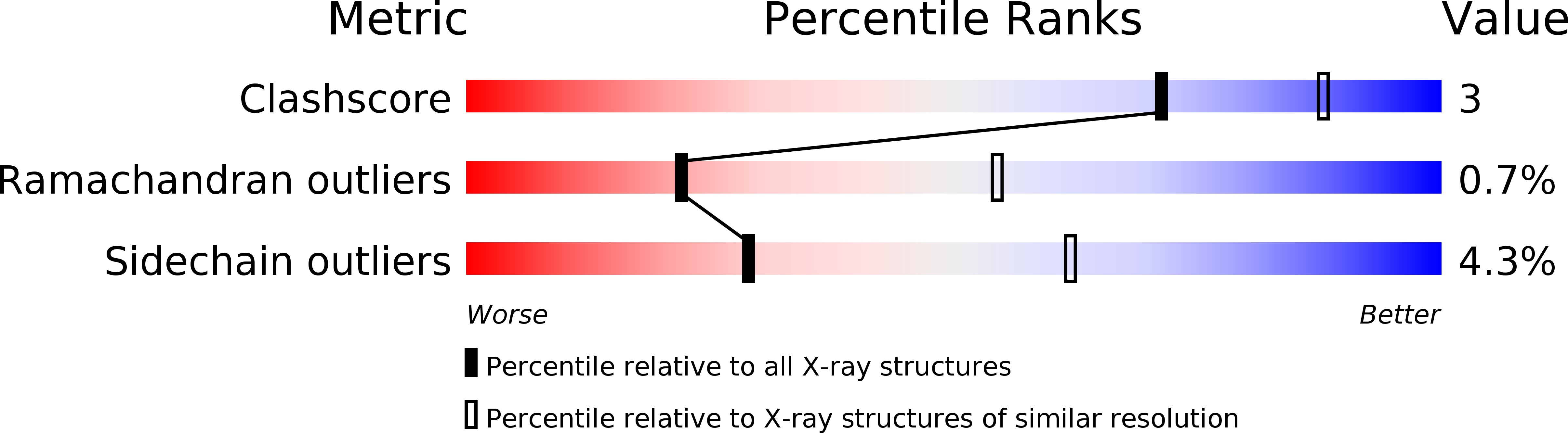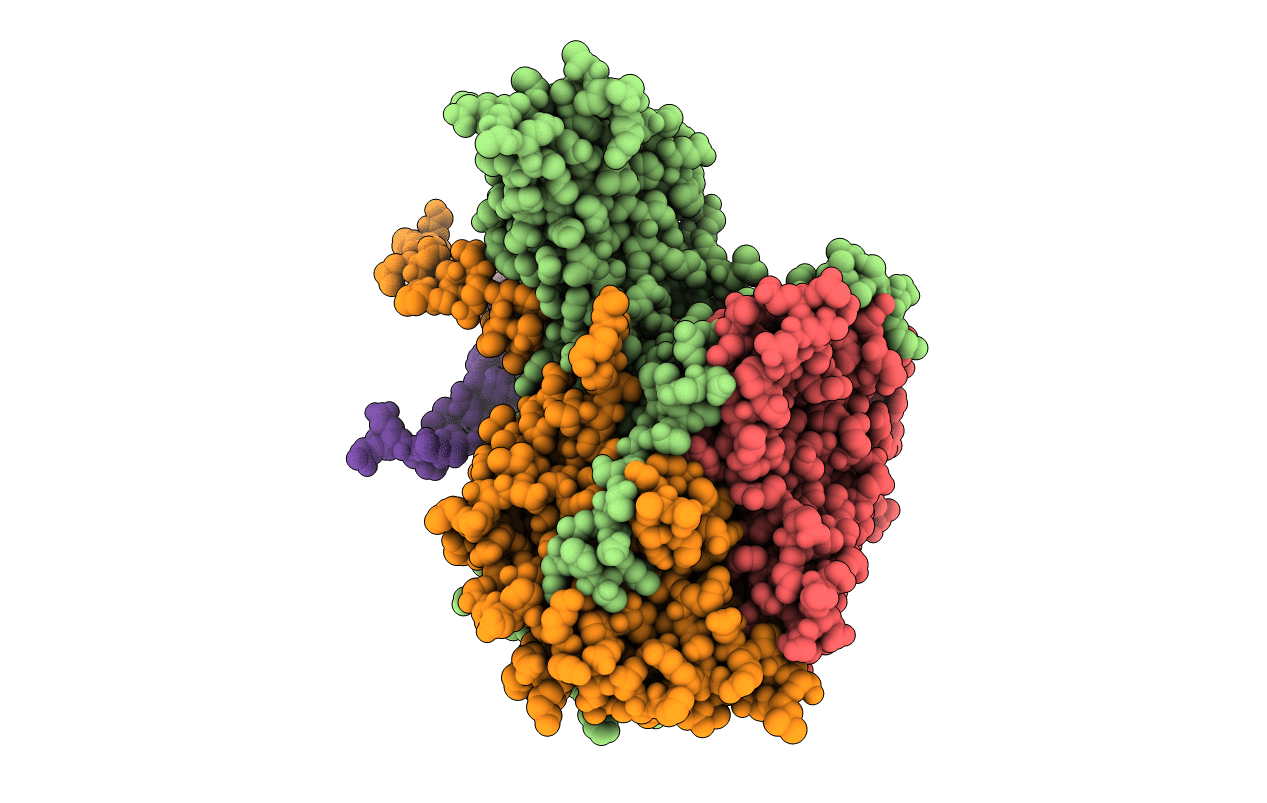
Deposition Date
1997-01-08
Release Date
1997-12-03
Last Version Date
2024-11-06
Entry Detail
PDB ID:
1PO2
Keywords:
Title:
POLIOVIRUS (TYPE 1, MAHONEY) IN COMPLEX WITH R77975, AN INHIBITOR OF VIRAL REPLICATION
Biological Source:
Source Organism(s):
Human poliovirus 1 (Taxon ID: 12081)
Method Details: