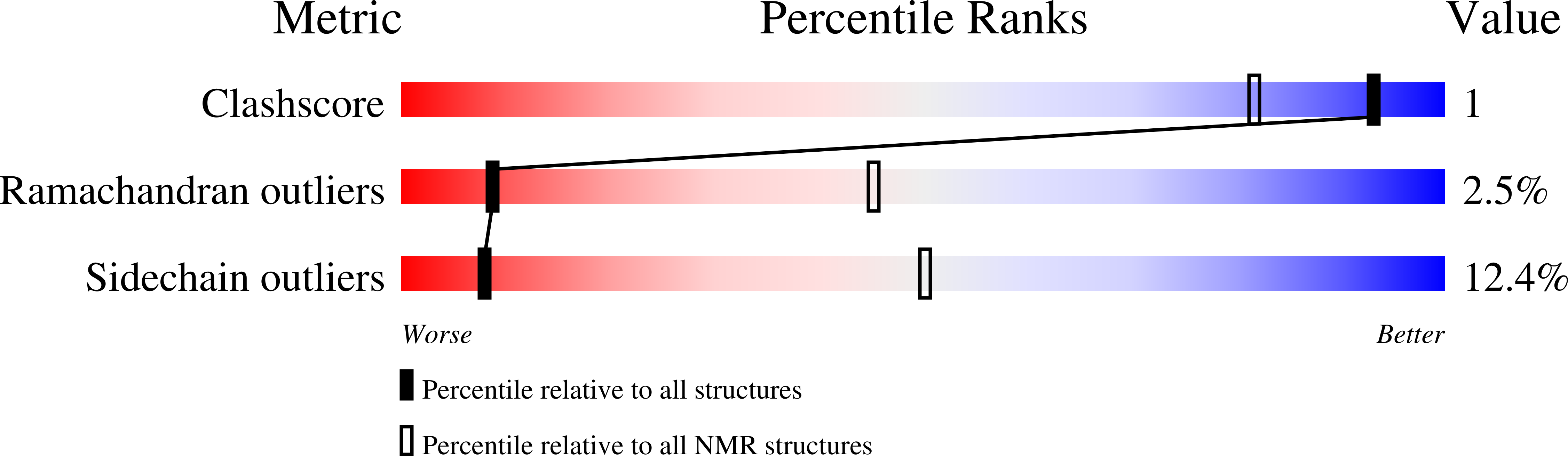
Deposition Date
1992-04-30
Release Date
1994-01-31
Last Version Date
2024-10-09
Entry Detail
PDB ID:
1PIT
Keywords:
Title:
DETERMINATION OF A HIGH-QUALITY NUCLEAR MAGNETIC RESONANCE SOLUTION STRUCTURE OF THE BOVINE PANCREATIC TRYPSIN INHIBITOR AND COMPARISON WITH THREE CRYSTAL STRUCTURES
Biological Source:
Source Organism:
Bos taurus (Taxon ID: 9913)
Method Details:
Experimental Method:
Conformers Submitted:
20