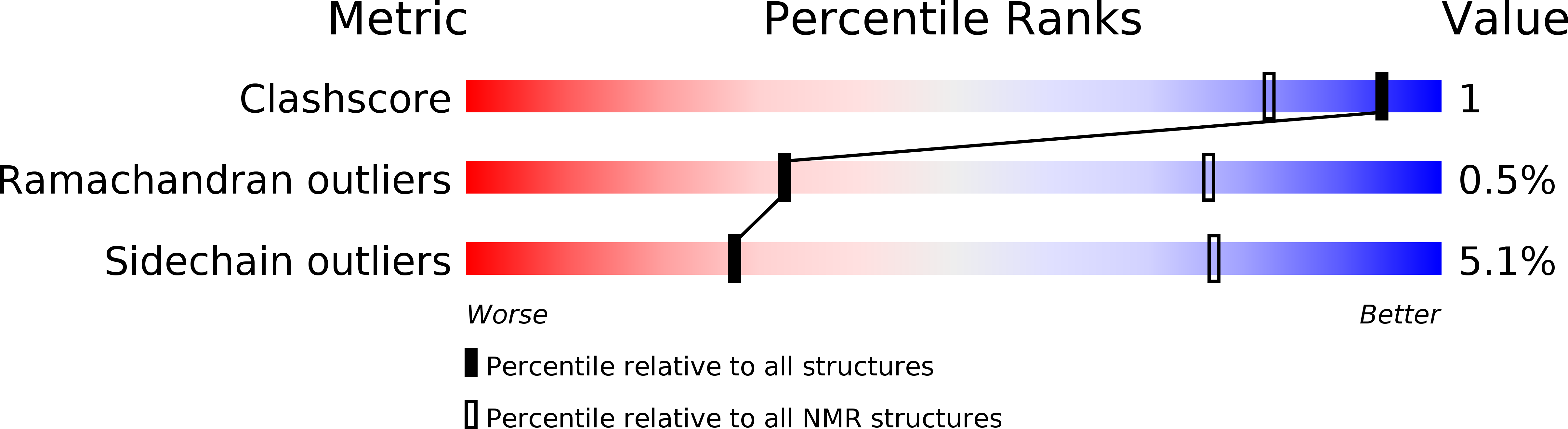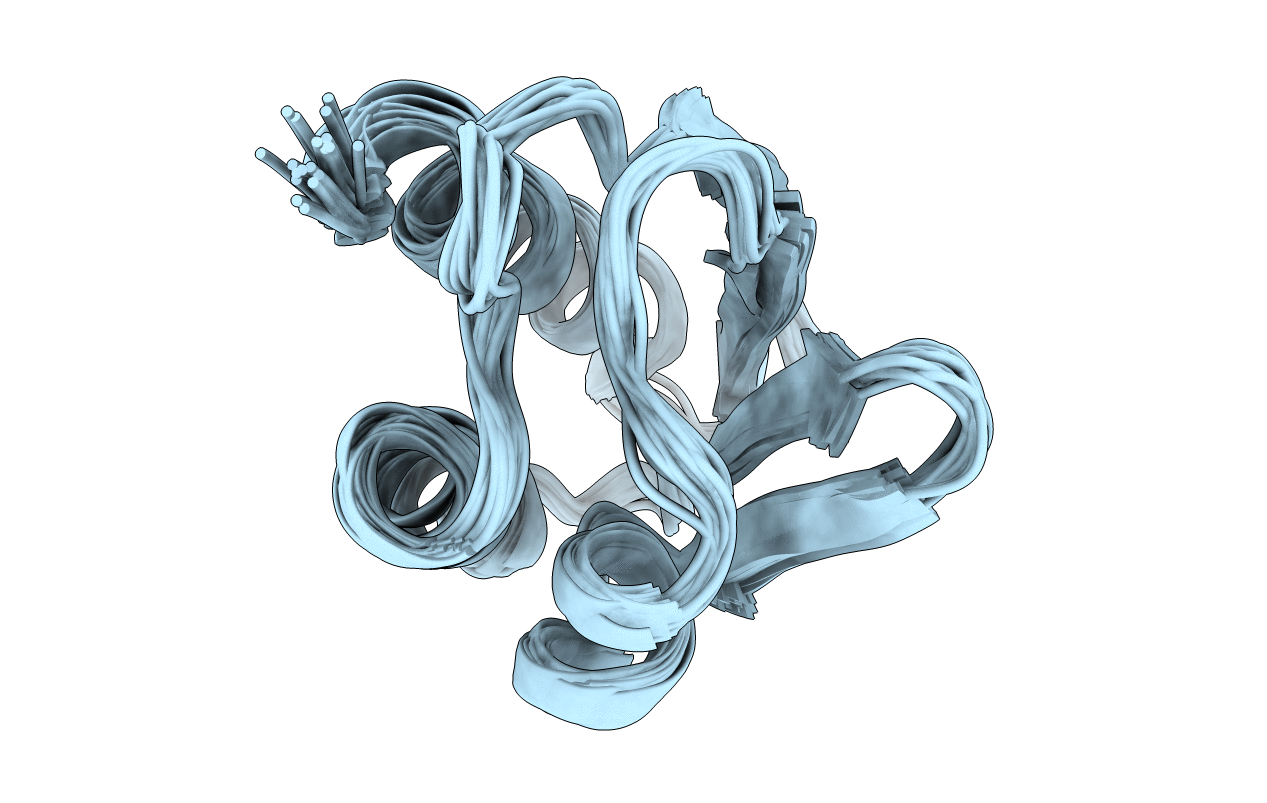
Deposition Date
1995-08-18
Release Date
1995-11-14
Last Version Date
2025-03-26
Entry Detail
PDB ID:
1PFH
Keywords:
Title:
THE PHOSPHORYLATED FORM OF THE HISTIDINE-CONTAINING PHOSPHOCARRIER PROTEIN HPR
Biological Source:
Source Organism(s):
Escherichia coli (Taxon ID: 562)
Method Details:
Experimental Method:
Conformers Submitted:
20