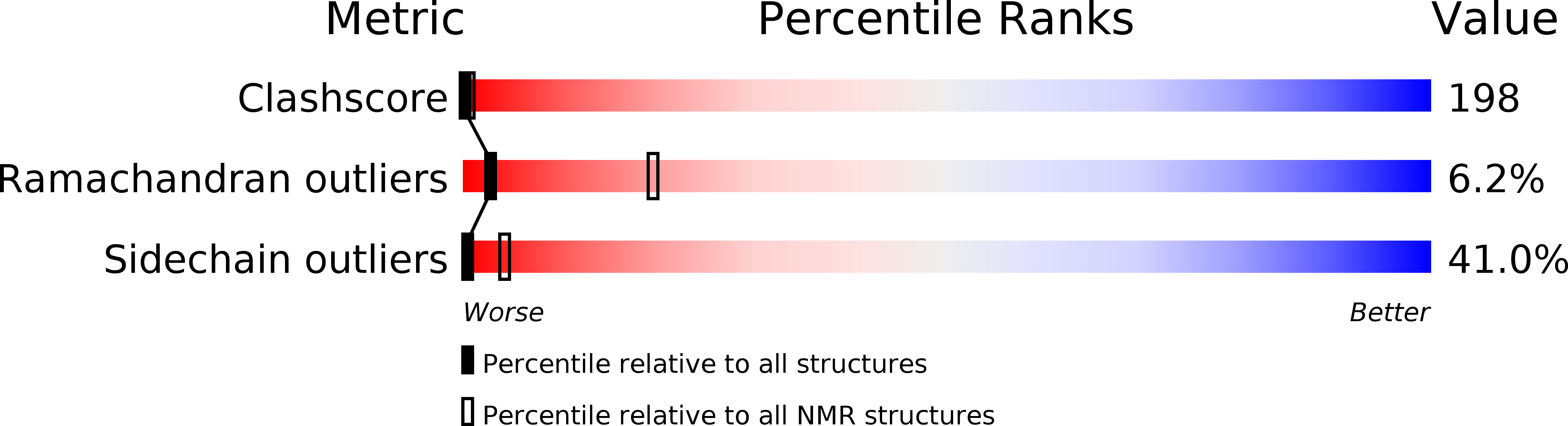
Deposition Date
1999-02-15
Release Date
1999-05-12
Last Version Date
2023-12-27
Entry Detail
Biological Source:
Source Organism(s):
Pseudomonas putida (Taxon ID: 303)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
80
Conformers Submitted:
14
Selection Criteria:
NO NOE VIOL.>0.5 A, NO DIHEDRAL VIOLATION > 5 DEG.