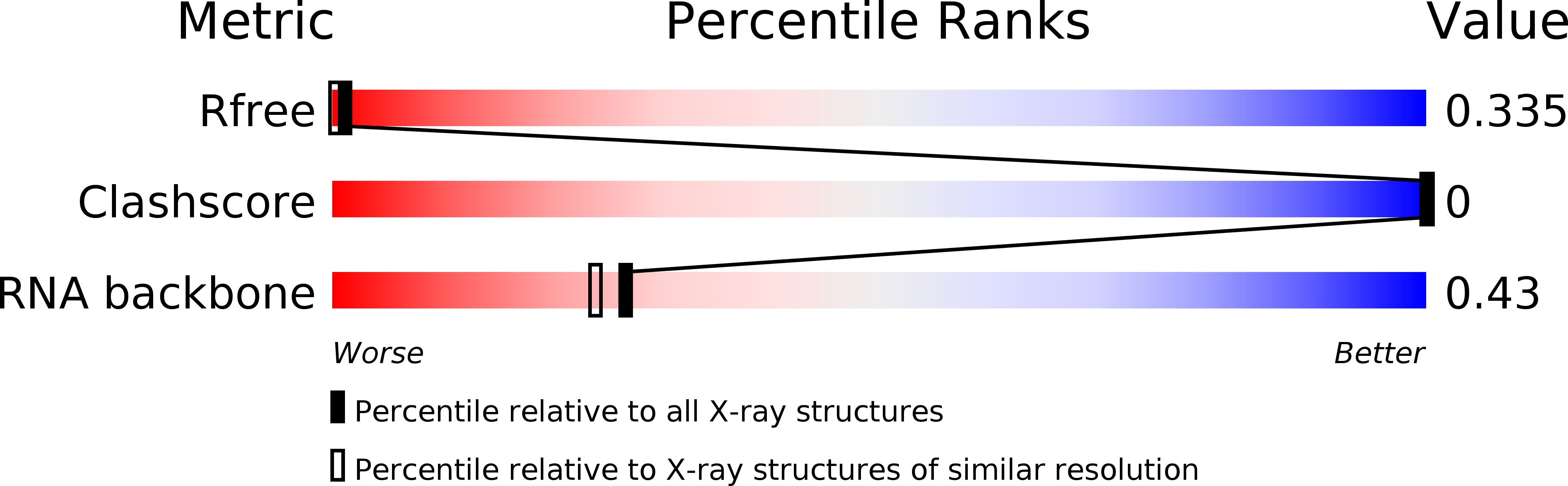Deposition Date
2003-04-30
Release Date
2003-11-04
Last Version Date
2024-02-14
Entry Detail
PDB ID:
1P79
Keywords:
Title:
Crystal structure of a bulged RNA tetraplex: implications for a novel binding site in RNA tetraplex
Method Details:
Experimental Method:
Resolution:
1.10 Å
R-Value Free:
0.13
R-Value Work:
0.12
R-Value Observed:
0.12
Space Group:
P 4 21 2