Deposition Date
2003-04-14
Release Date
2004-06-01
Last Version Date
2024-02-14
Entry Detail
PDB ID:
1P1X
Keywords:
Title:
Comparison of class I aldolase binding site architecture based on the crystal structure of 2-deoxyribose-5-phosphate aldolase determined at 0.99 Angstrom resolution
Biological Source:
Source Organism(s):
Escherichia coli K-12 (Taxon ID: 83333)
Expression System(s):
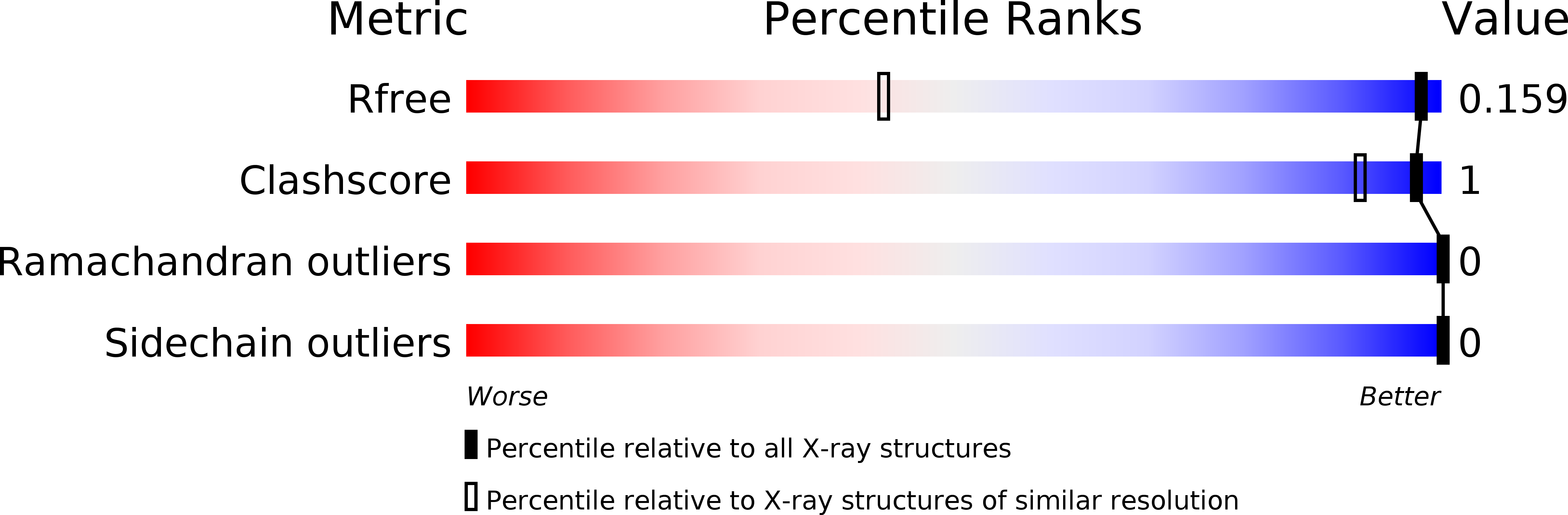
Method Details: