Deposition Date
2003-04-02
Release Date
2004-04-06
Last Version Date
2024-10-09
Entry Detail
PDB ID:
1OXL
Keywords:
Title:
INHIBITION OF PHOSPHOLIPASE A2 (PLA2) BY (2-CARBAMOYLMETHYL-5-PROPYL-OCTAHYDRO-INDOL-7-YL)-ACETIC ACID (INDOLE): CRYSTAL STRUCTURE OF THE COMPLEX FORMED BETWEEN PLA2 FROM RUSSELL'S VIPER AND INDOLE AT 1.8 RESOLUTION
Biological Source:
Source Organism:
Daboia russellii russellii (Taxon ID: 31159)
Method Details:
Experimental Method:
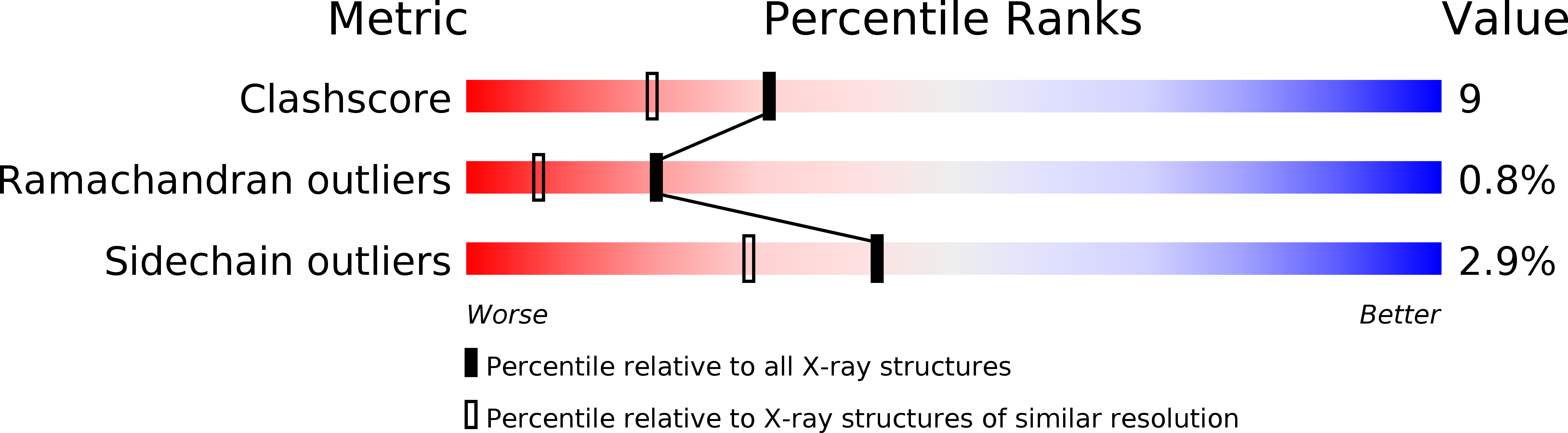
Resolution:
1.80 Å
R-Value Free:
0.23
R-Value Work:
0.18
R-Value Observed:
0.19
Space Group:
C 2 2 21