Deposition Date
2003-07-10
Release Date
2004-01-07
Last Version Date
2024-11-06
Entry Detail
PDB ID:
1OJI
Keywords:
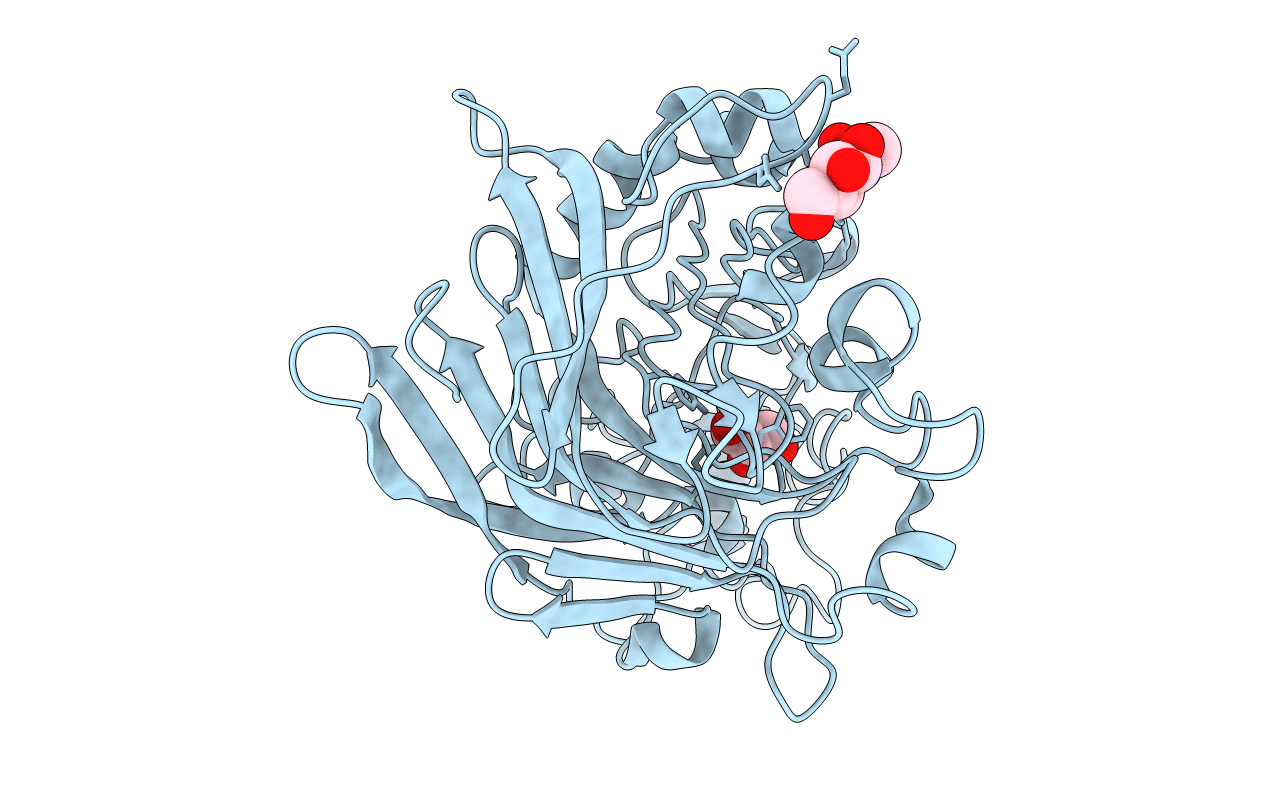
Title:
Anatomy of glycosynthesis: Structure and kinetics of the Humicola insolens Cel7B E197A and E197S glycosynthase mutants
Biological Source:
Source Organism:
HUMICOLA INSOLENS (Taxon ID: 34413)
Host Organism:
Method Details:
Experimental Method:
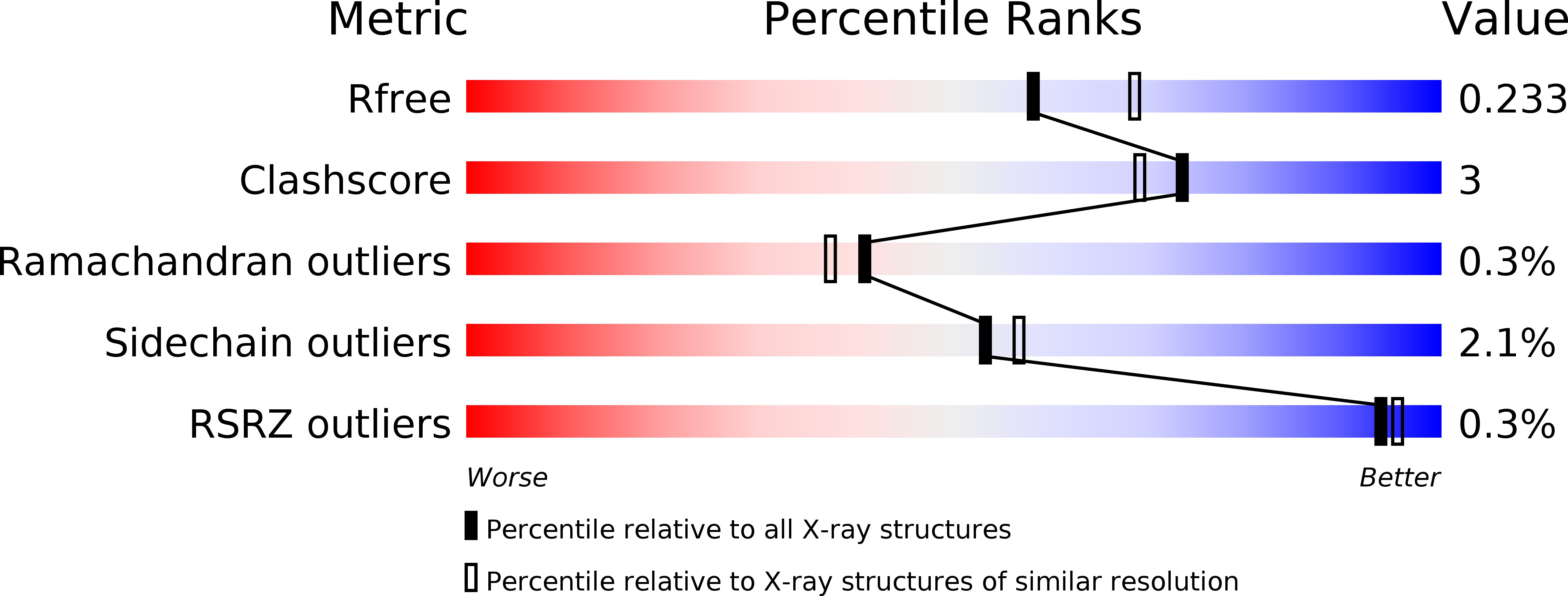
Resolution:
2.15 Å
R-Value Free:
0.22
R-Value Work:
0.19
R-Value Observed:
0.19
Space Group:
P 65