Deposition Date
2003-05-08
Release Date
2003-10-09
Last Version Date
2023-12-13
Entry Detail
PDB ID:
1OGP
Keywords:
Title:
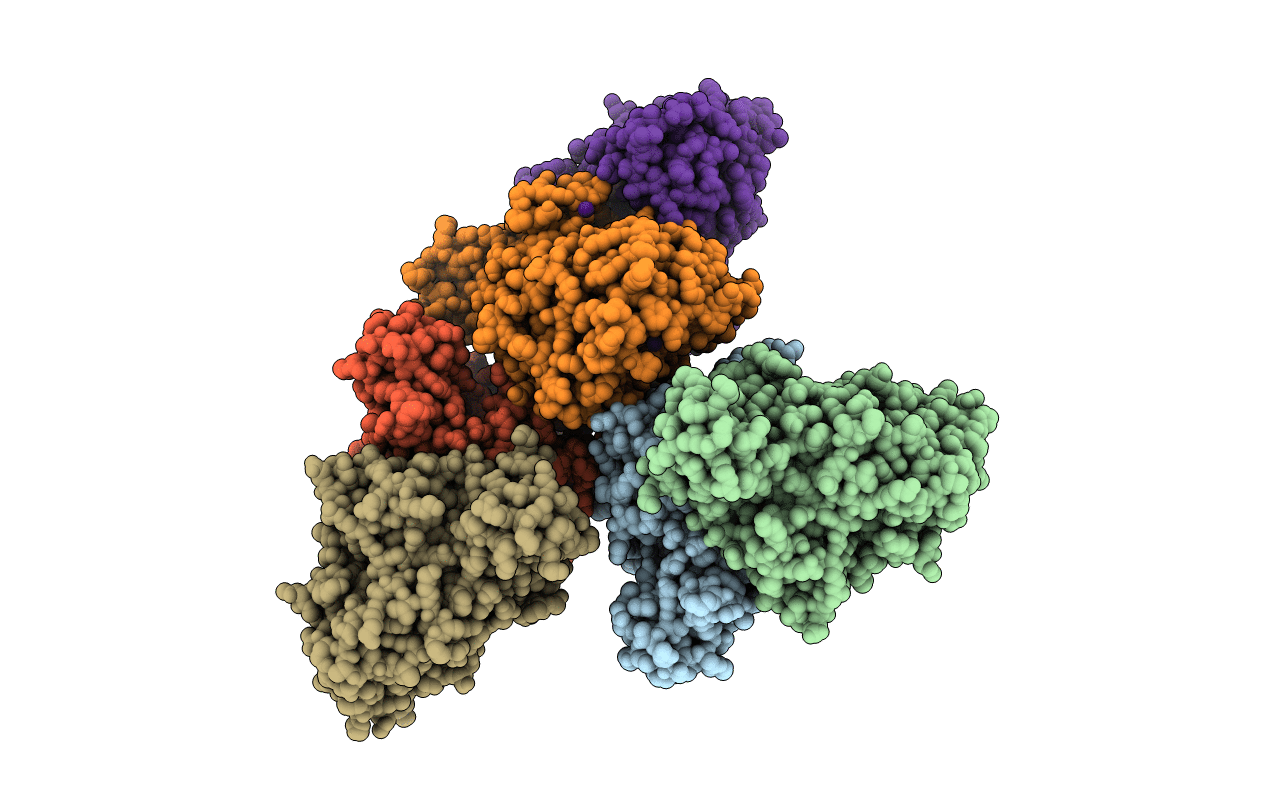
The crystal structure of plant sulfite oxidase provides insight into sulfite oxidation in plants and animals
Biological Source:
Source Organism(s):
ARABIDOPSIS THALIANA (Taxon ID: 3702)
Expression System(s):
Method Details:
Experimental Method:
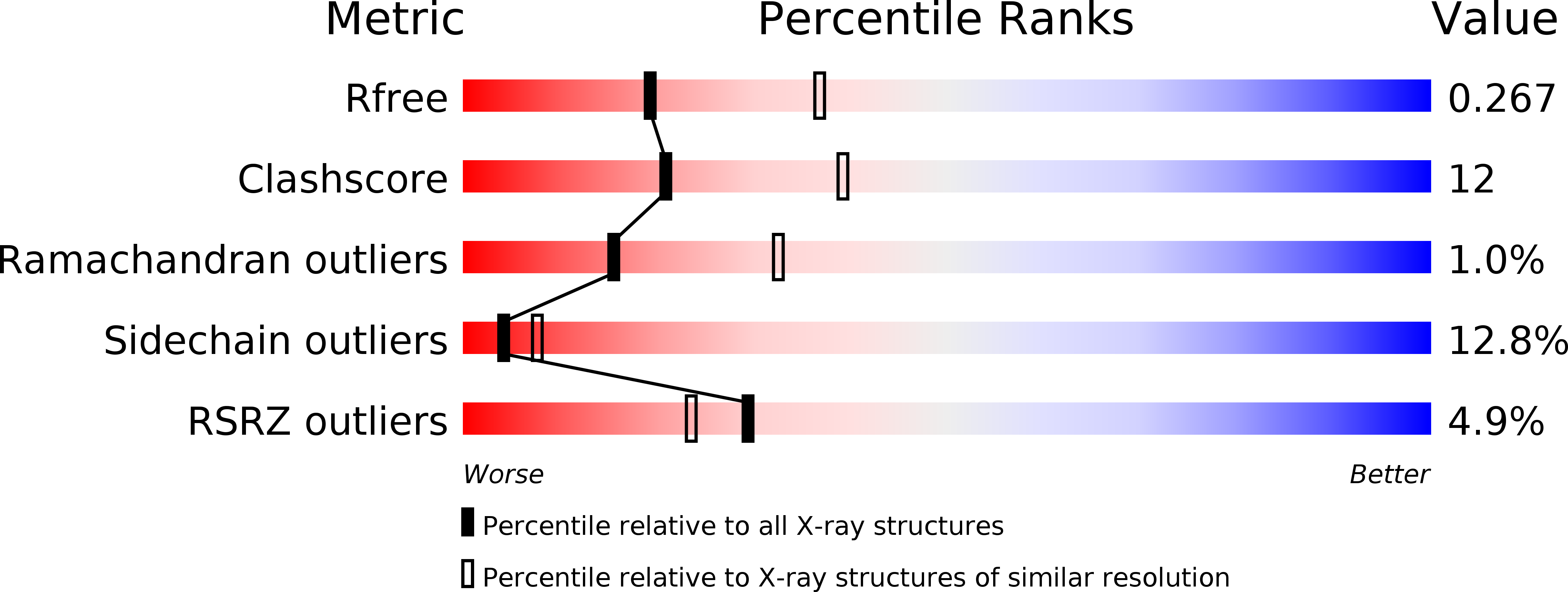
Resolution:
2.60 Å
R-Value Free:
0.26
R-Value Work:
0.23
R-Value Observed:
0.23
Space Group:
C 2 2 2