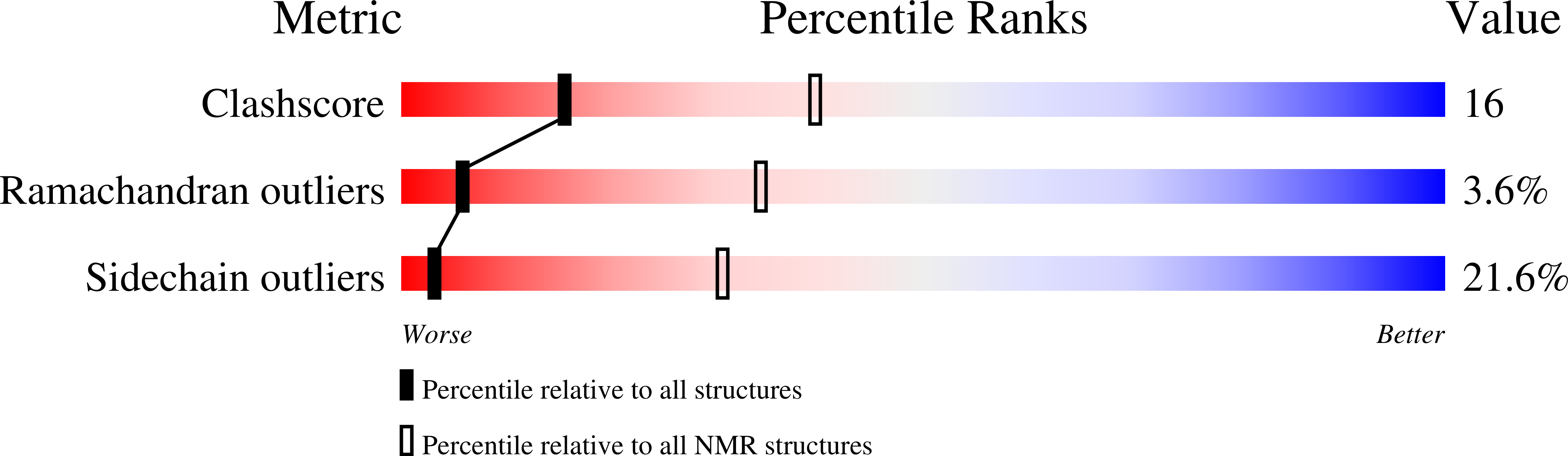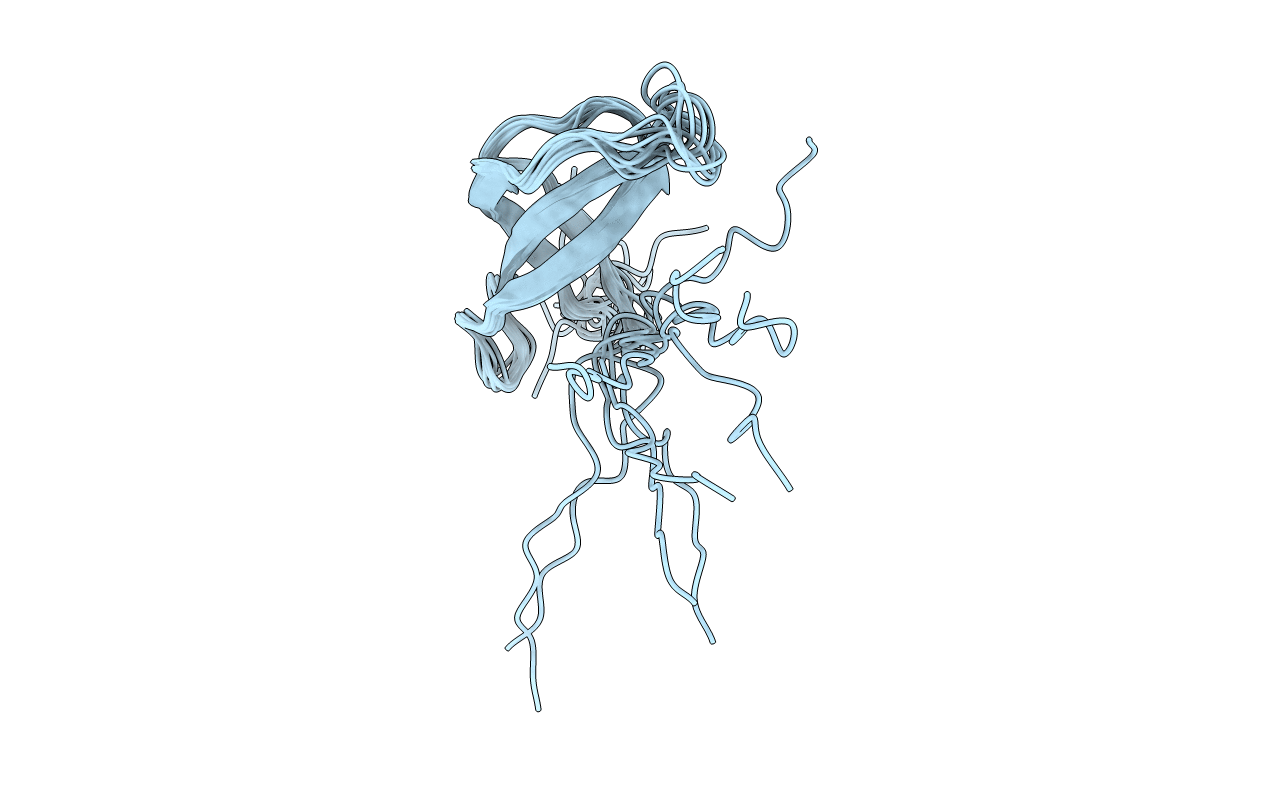
Deposition Date
2003-02-11
Release Date
2003-09-02
Last Version Date
2024-05-22
Entry Detail
PDB ID:
1NY4
Keywords:
Title:
Solution structure of the 30S ribosomal protein S28E from Pyrococcus horikoshii. Northeast Structural Genomics Consortium target JR19.
Biological Source:
Source Organism:
Pyrococcus horikoshii (Taxon ID: 53953)
Host Organism:
Method Details:
Experimental Method:
Conformers Calculated:
56
Conformers Submitted:
10
Selection Criteria:
target function