Abstact
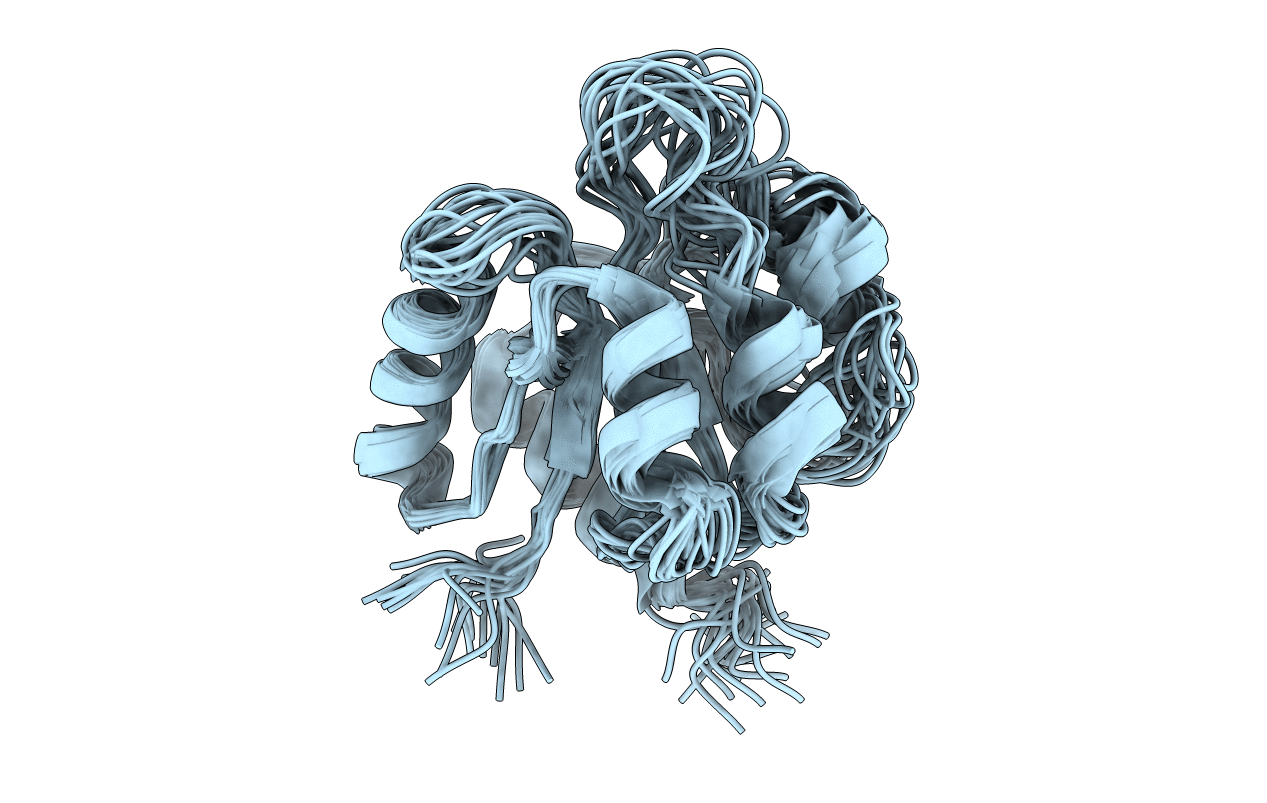
NTRC is a transcriptional enhancer binding protein whose N-terminal domain is a member of the family of receiver domains of two-component regulatory systems. Using 3D and 4D NMR spectroscopy, we have completed the 1H, 15N, and 13C assignments and determined the solution structure of the N-terminal receiver domain of the NTRC protein. Determination of the three-dimensional structure was carried out with the program X-PLOR (Brünger, 1992) using a total of 915 NMR-derived distance and dihedral angle restraints. The resultant family of structures has an average root mean square deviation of 0.81 A from the average structure for the backbone atoms involved in well-defined secondary structure. The structure is comprised of five alpha-helices and a five-stranded parallel beta-sheet, in a (beta/alpha)5 topology. Comparison of the solution structure of the NTRC receiver domain with the crystal structures of the homologous protein CheY in both the Mg(2+)-free and Mg(2+)-bound forms [Stock, A.M., Mottonen, J. M., Stock, J. B., & Schutt, C. E. (1989) Nature 337, 745-749; Volz, K., & Matsumura, P. (1991) J. Biol. Chem. 296, 15511-15519; Stock, A. M., Martinez-Hackert, E., Rasmussen, B. F., West, A. H., Stock, J. B., Ringe, D., & Petsko, G. A. (1993) Biochemistry 32, 13375-13380; Bellsolell, L., Prieto, J., Serrano, L., & Coll, M. (1994) J. Mol. Biol. 238, 489-495] reveals a very similar fold, with the only significant difference occurring in the positioning of helix 4 relative to the rest of the protein. Examination of the conformation of consensus residues of the receiver domain superfamily [Volz, K. (1993) Biochemistry 32, 11741-11753] in the structures of the NTRC receiver domain and CheY establishes the structural importance of residues whose side chains are involved in hydrogen bonding or hydrophobic core interactions. The importance of some nonconsensus residues which may be conserved for their ability to fulfill helix capping roles is also discussed.