Deposition Date
1996-02-05
Release Date
1996-07-11
Last Version Date
2024-05-01
Entry Detail
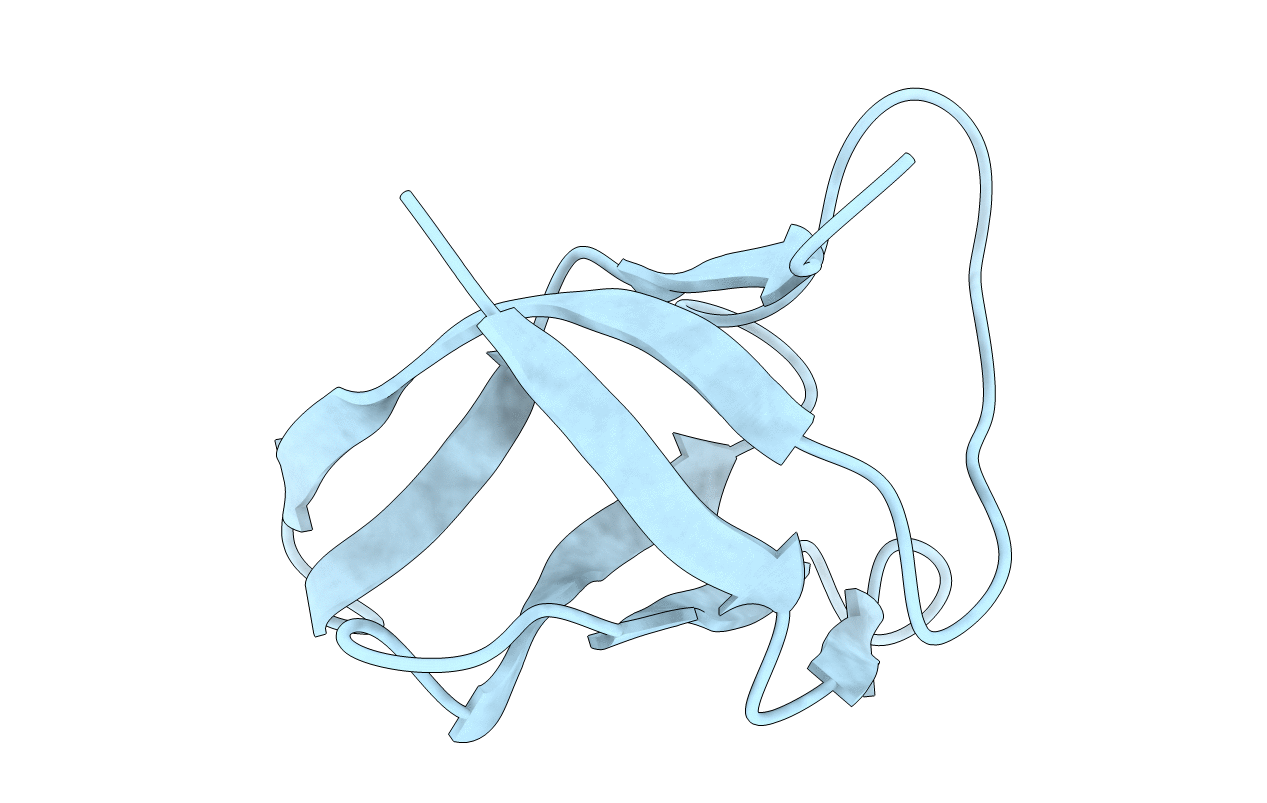
PDB ID:
1NMG
Keywords:
Title:
MAJOR COLD-SHOCK PROTEIN, NMR, MINIMIZED AVERAGE STRUCTURE
Biological Source:
Source Organism(s):
Bacillus subtilis (Taxon ID: 1423)
Expression System(s):
Method Details:
Experimental Method:
Conformers Submitted:
1


