Deposition Date
2002-12-31
Release Date
2003-08-26
Last Version Date
2023-08-16
Entry Detail
PDB ID:
1NJJ
Keywords:
Title:
Crystal structure determination of T. brucei ornithine decarboxylase bound to D-ornithine and to G418
Biological Source:
Source Organism:
Trypanosoma brucei gambiense (Taxon ID: 31285)
Host Organism:
Method Details:
Experimental Method:
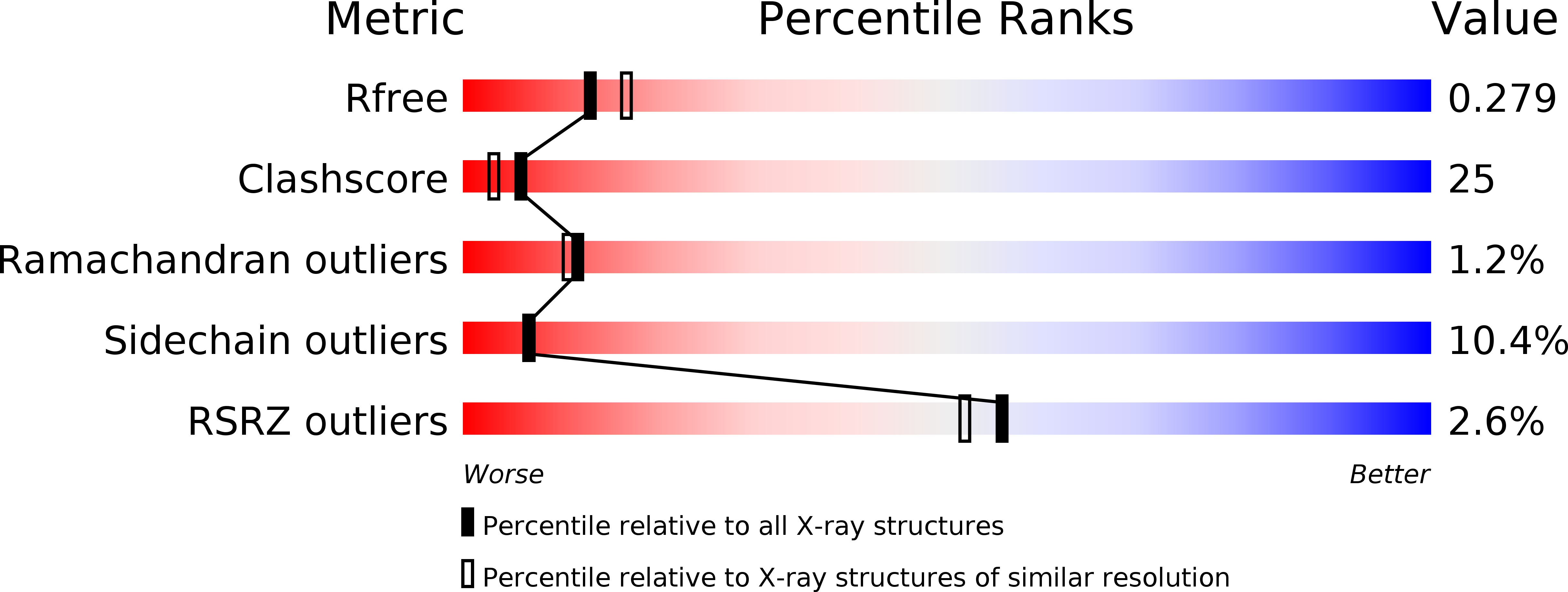
Resolution:
2.45 Å
R-Value Free:
0.28
R-Value Work:
0.26
Space Group:
P 1 21 1