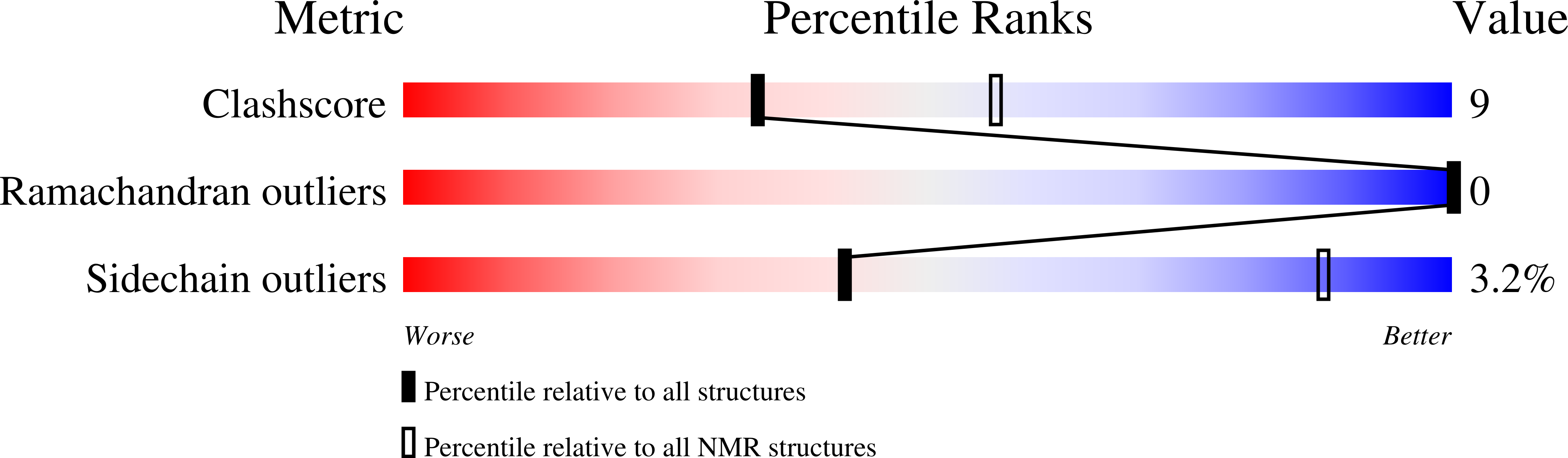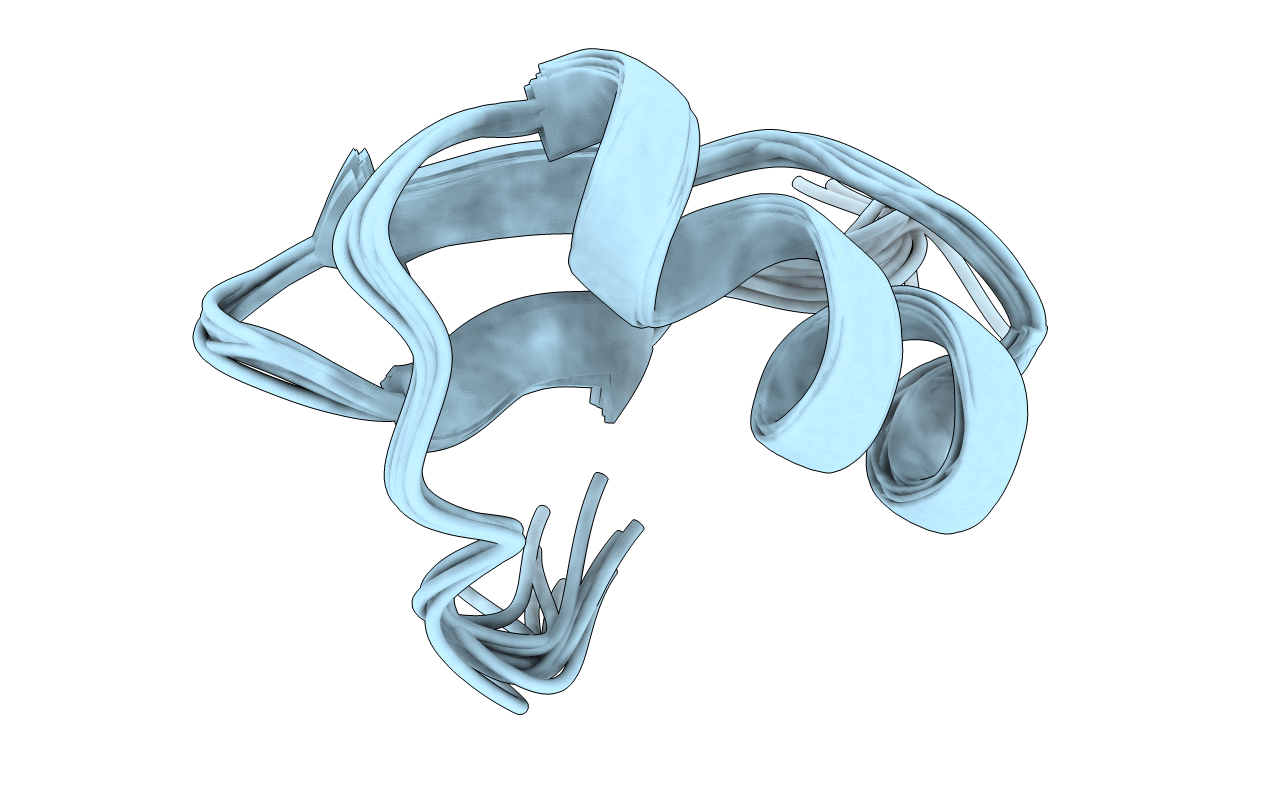
Deposition Date
2002-11-21
Release Date
2003-09-02
Last Version Date
2024-11-06
Entry Detail
PDB ID:
1N8M
Keywords:
Title:
Solution structure of Pi4, a four disulfide bridged scorpion toxin active on potassium channels
Method Details:
Experimental Method:
Conformers Calculated:
20
Conformers Submitted:
10
Selection Criteria:
structures with the lowest energy