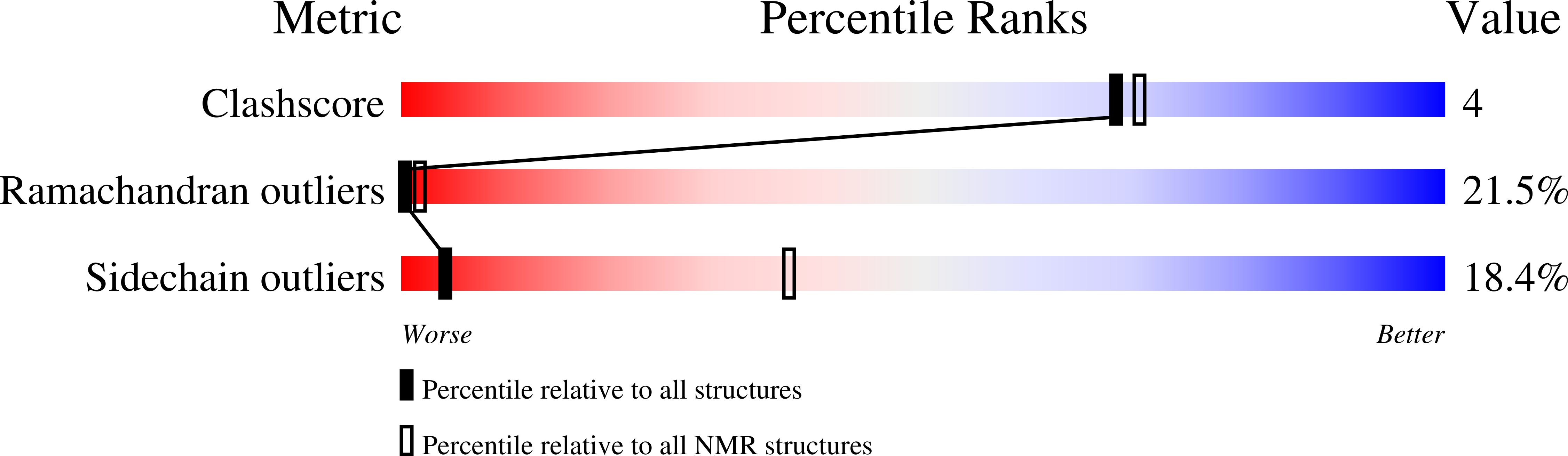
Deposition Date
2002-10-08
Release Date
2003-09-02
Last Version Date
2024-05-29
Entry Detail
PDB ID:
1MZI
Keywords:
Title:
Solution ensemble structures of HIV-1 gp41 2F5 mAb epitope
Method Details:
Experimental Method:
Conformers Calculated:
6400
Conformers Submitted:
81
Selection Criteria:
Base set ensemble representative of the conformational space experimentally allowed