Deposition Date
2002-09-26
Release Date
2003-04-01
Last Version Date
2024-02-14
Entry Detail
PDB ID:
1MVR
Keywords:
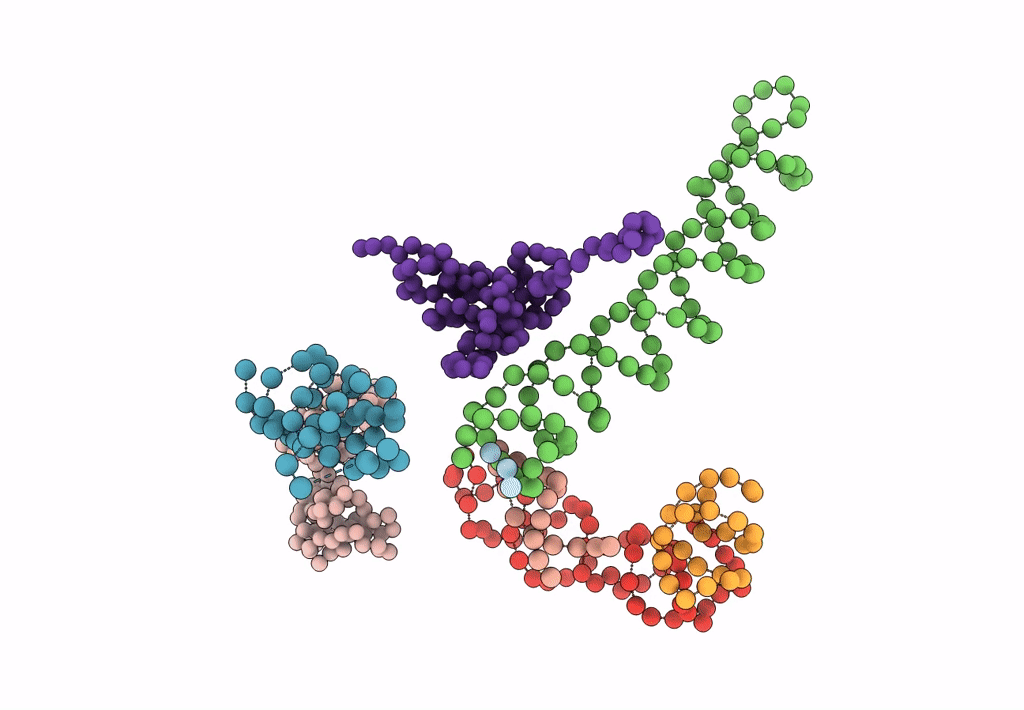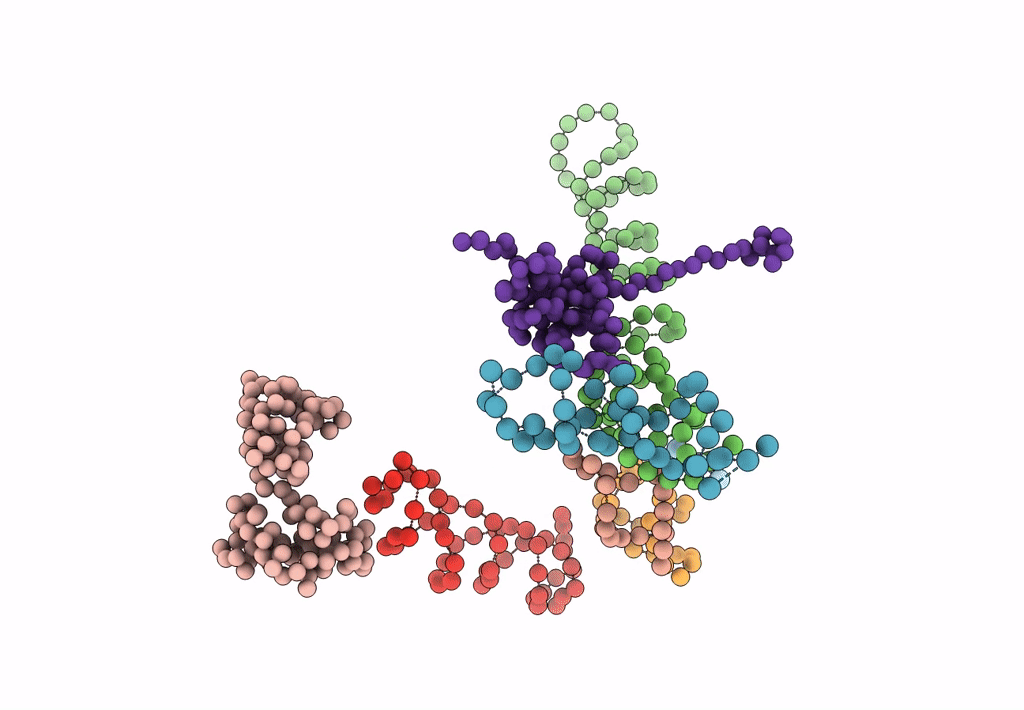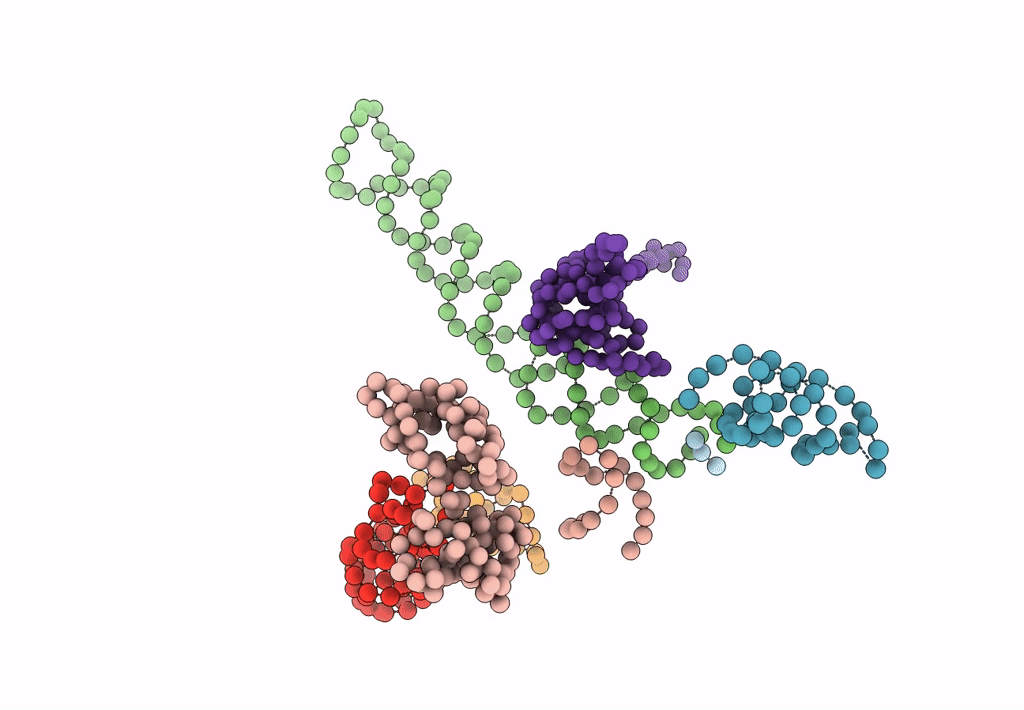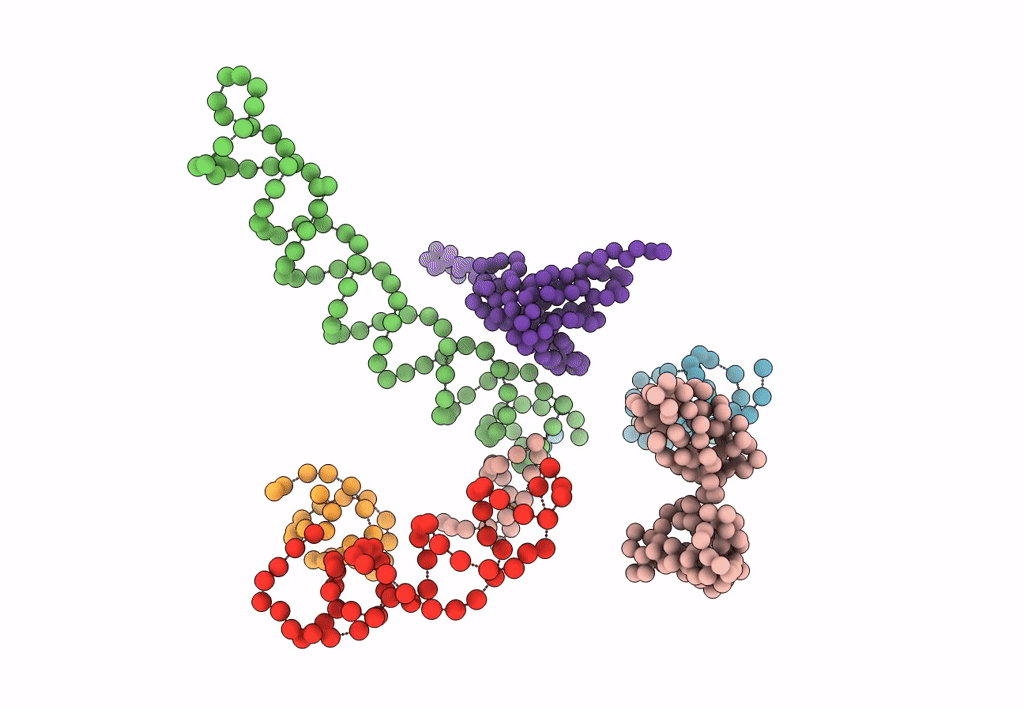
Title:
Decoding Center & Peptidyl transferase center from the X-ray structure of the Thermus thermophilus 70S ribosome, aligned to the low resolution Cryo-EM map of E.coli 70S Ribosome
Biological Source:
Source Organism(s):
Escherichia coli (Taxon ID: 83333)
Method Details:
Experimental Method:
Resolution:
12.80 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE


