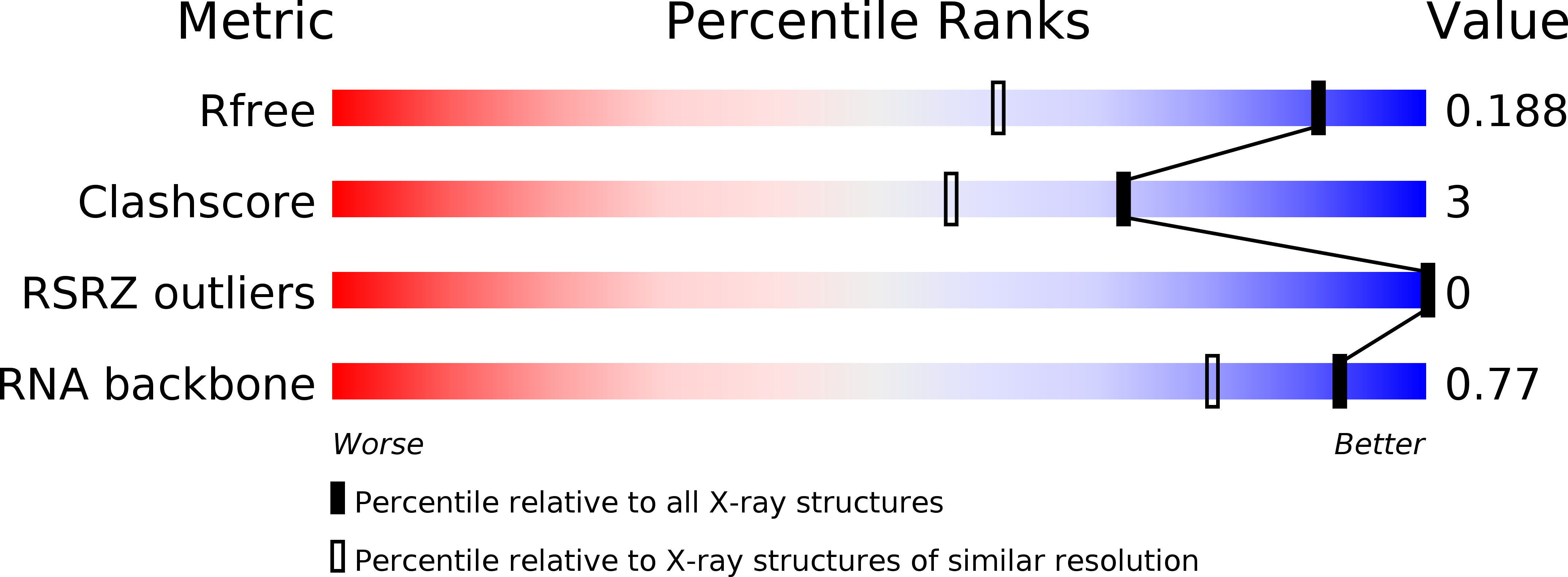
Deposition Date
2002-09-19
Release Date
2003-02-25
Last Version Date
2024-02-14
Entry Detail
PDB ID:
1MSY
Keywords:
Title:
GUAA tetraloop mutant of Sarcin/Ricin domain from E. Coli 23 S rRNA
Method Details:
Experimental Method:
Resolution:
1.41 Å
R-Value Free:
0.20
R-Value Work:
0.16
R-Value Observed:
0.16
Space Group:
C 1 2 1