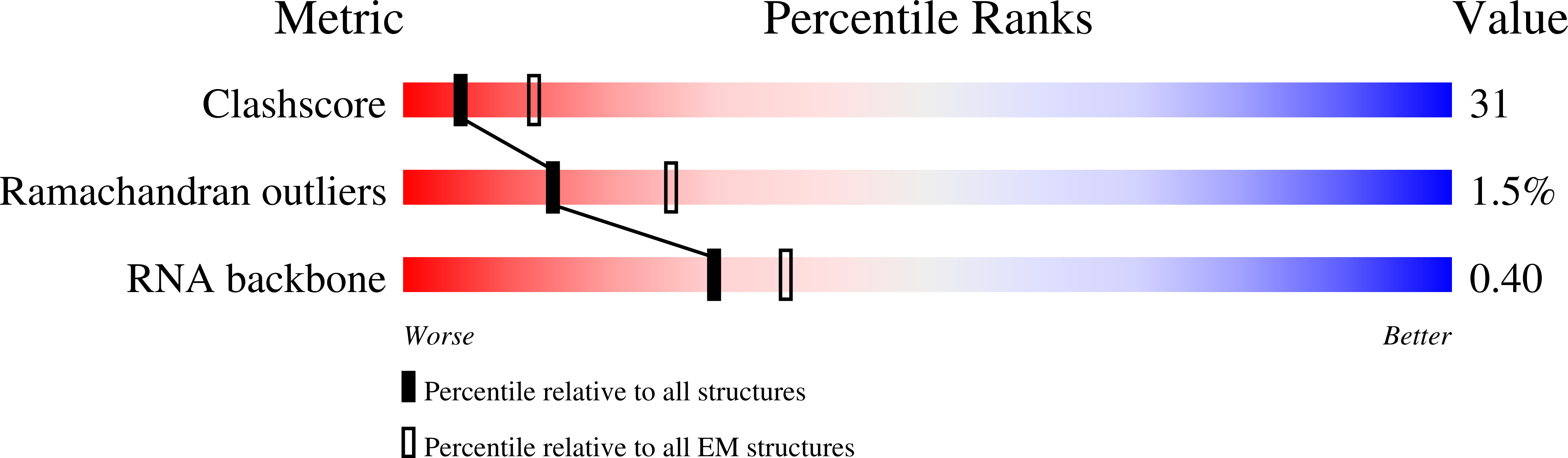
Deposition Date
2002-08-26
Release Date
2002-11-01
Last Version Date
2024-02-14
Entry Detail
PDB ID:
1MJ1
Keywords:
Title:
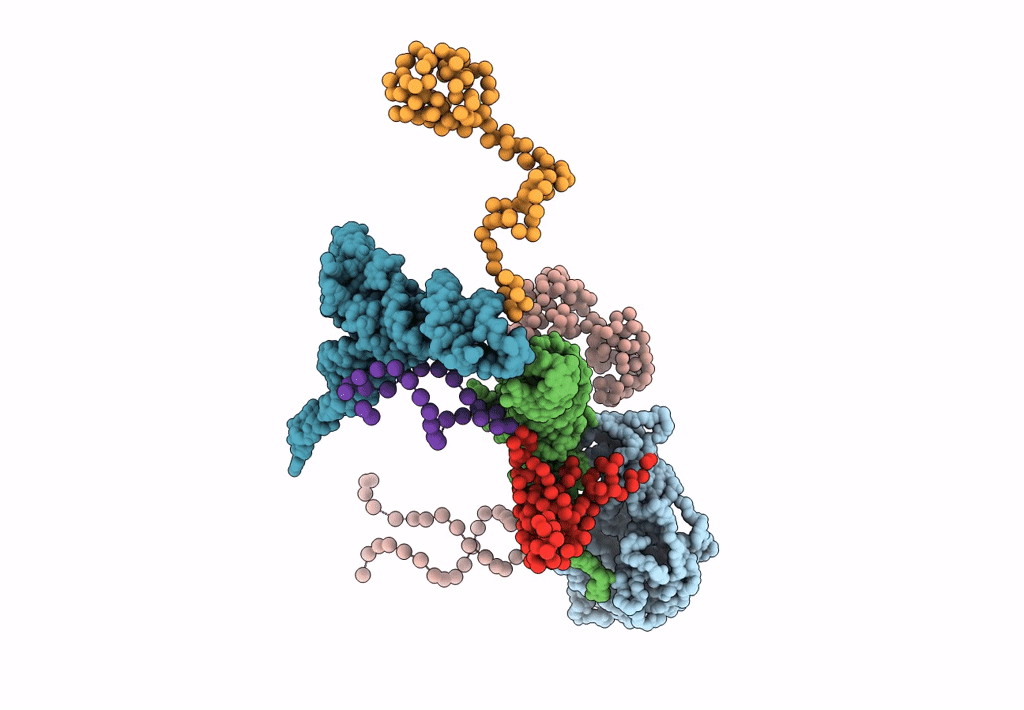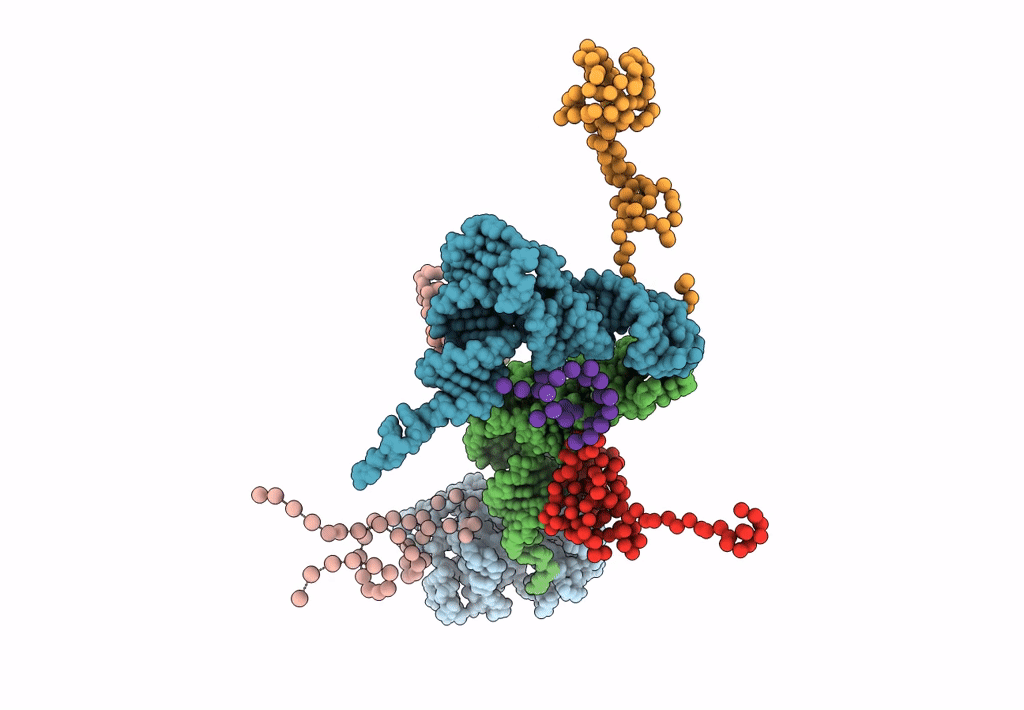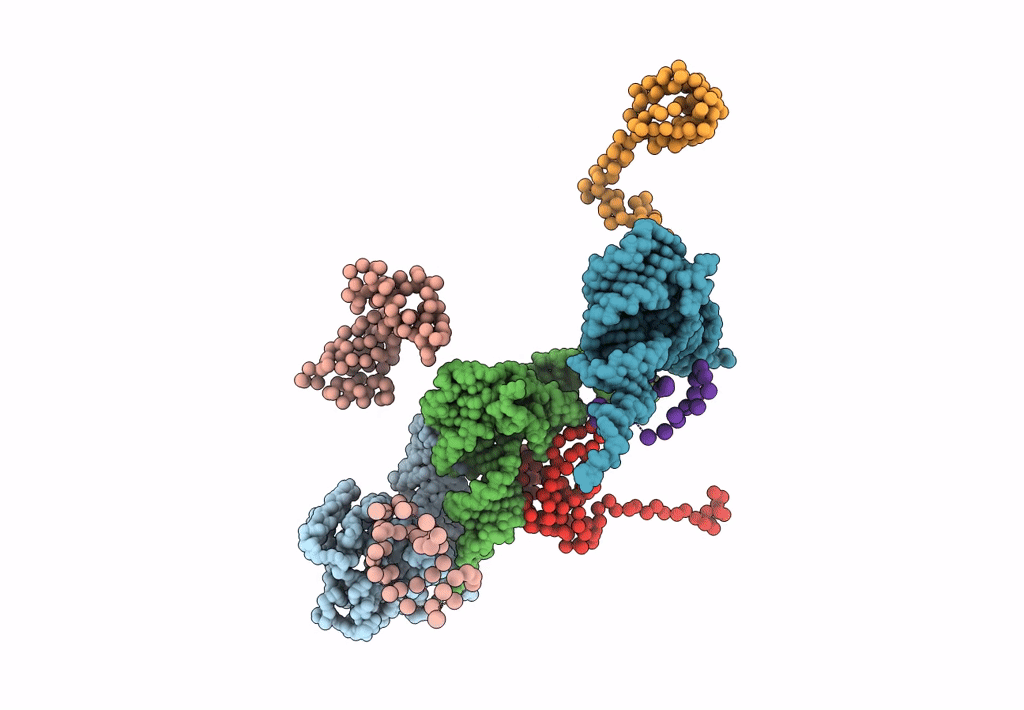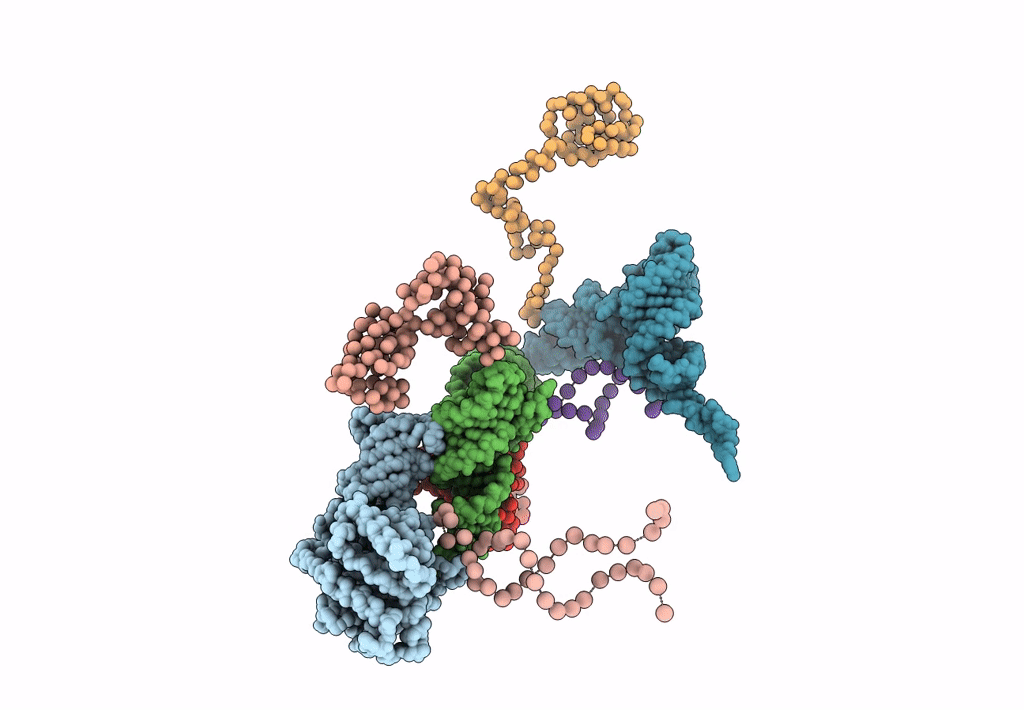
FITTING THE TERNARY COMPLEX OF EF-Tu/tRNA/GTP AND RIBOSOMAL PROTEINS INTO A 13 A CRYO-EM MAP OF THE COLI 70S RIBOSOME
Biological Source:
Source Organism(s):
Escherichia coli (Taxon ID: 562)
Method Details:
Experimental Method:
Resolution:
13.00 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE